teashirt zinc finger homeobox 2
ZFIN 
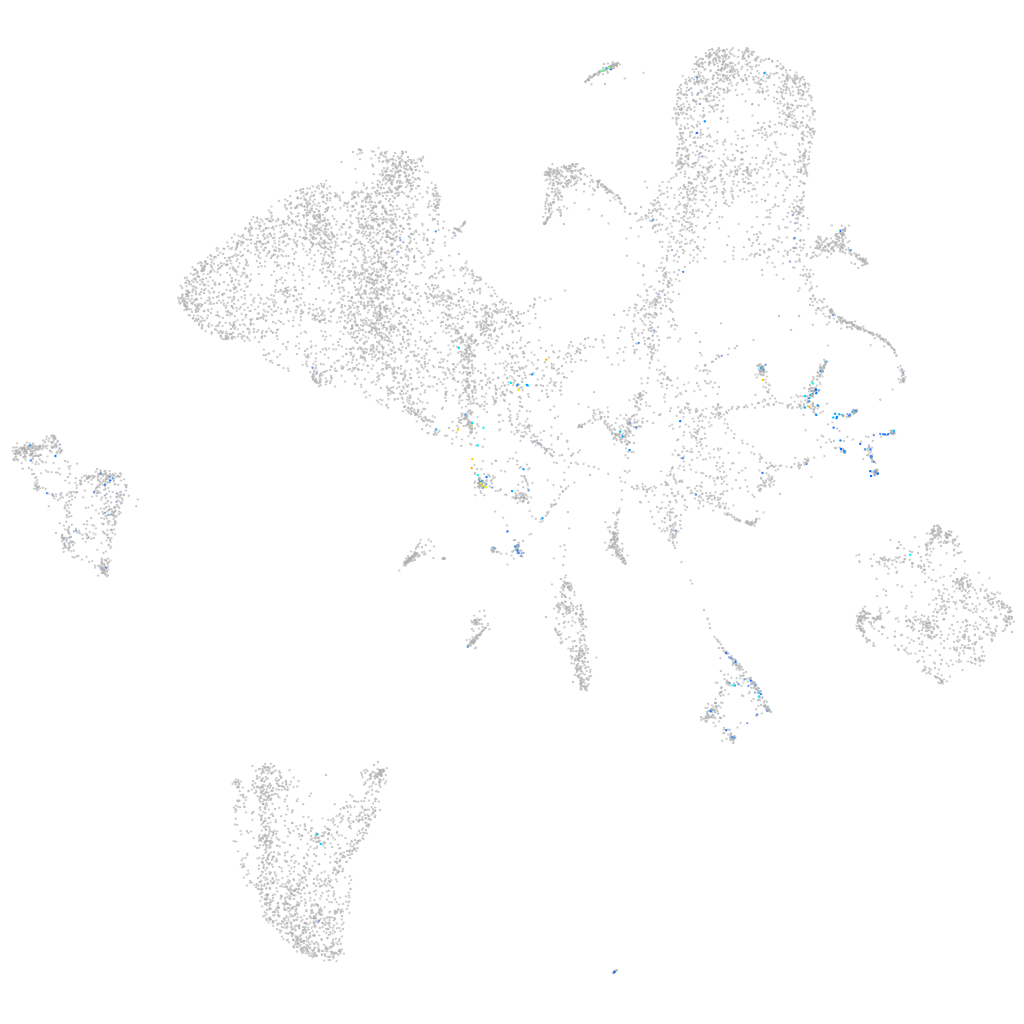
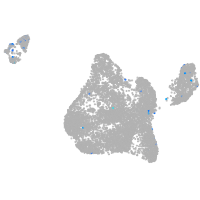
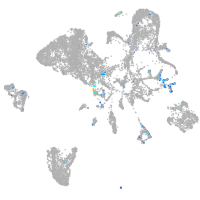
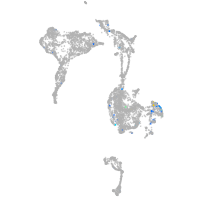
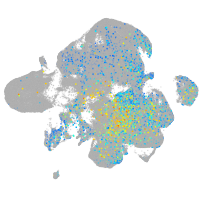
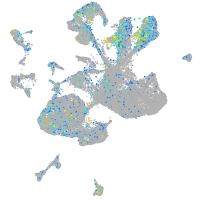
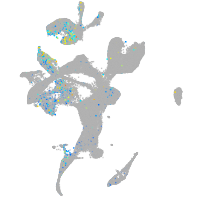
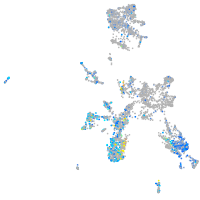
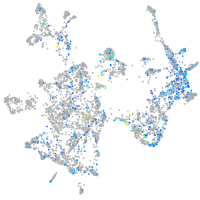
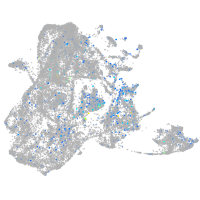
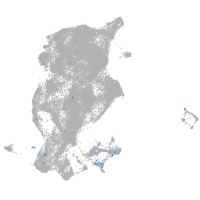
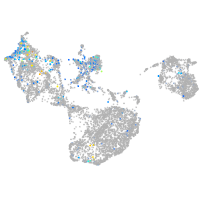
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod1 | 0.202 | gapdh | -0.107 |
| mtus2a | 0.195 | ahcy | -0.106 |
| scgn | 0.193 | aldob | -0.099 |
| gnao1a | 0.192 | gatm | -0.099 |
| stmn1b | 0.192 | hdlbpa | -0.096 |
| cacng8b | 0.188 | gamt | -0.094 |
| rtn1b | 0.186 | fbp1b | -0.092 |
| nr4a2a | 0.184 | bhmt | -0.091 |
| gpc1a | 0.179 | mat1a | -0.091 |
| si:dkey-12h9.6 | 0.178 | aqp12 | -0.087 |
| tuba1c | 0.177 | gstt1a | -0.086 |
| gpr85 | 0.175 | gstr | -0.085 |
| LOC101883913 | 0.173 | gpx4a | -0.084 |
| isl1 | 0.171 | aldh7a1 | -0.082 |
| pax6b | 0.171 | scp2a | -0.082 |
| arxa | 0.171 | agxtb | -0.081 |
| gapdhs | 0.170 | apoa4b.1 | -0.081 |
| atp1a3a | 0.169 | aldh6a1 | -0.081 |
| LOC100537384 | 0.169 | gnmt | -0.080 |
| ptprsb | 0.168 | hspe1 | -0.080 |
| slc35g2b | 0.166 | dap | -0.080 |
| pcsk2 | 0.166 | cx32.3 | -0.079 |
| vat1 | 0.165 | eno3 | -0.078 |
| kcnj11 | 0.164 | slc37a4a | -0.078 |
| adam22 | 0.163 | nipsnap3a | -0.078 |
| nsg2 | 0.162 | apoc2 | -0.076 |
| chd3 | 0.162 | apoa1b | -0.076 |
| tmem178b | 0.162 | tdo2a | -0.076 |
| tmsb | 0.162 | afp4 | -0.075 |
| pcdh2ab7 | 0.158 | glud1b | -0.075 |
| LOC101882396 | 0.158 | g6pca.2 | -0.075 |
| gpm6aa | 0.158 | rdh1 | -0.075 |
| elavl3 | 0.158 | fdx1 | -0.075 |
| rims2a | 0.157 | apoc1 | -0.075 |
| scg3 | 0.157 | dcxr | -0.074 |