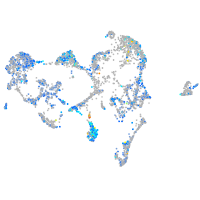
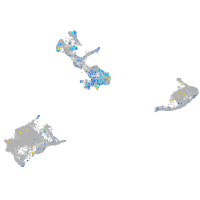
"TSC22 domain family, member 1"
ZFIN 
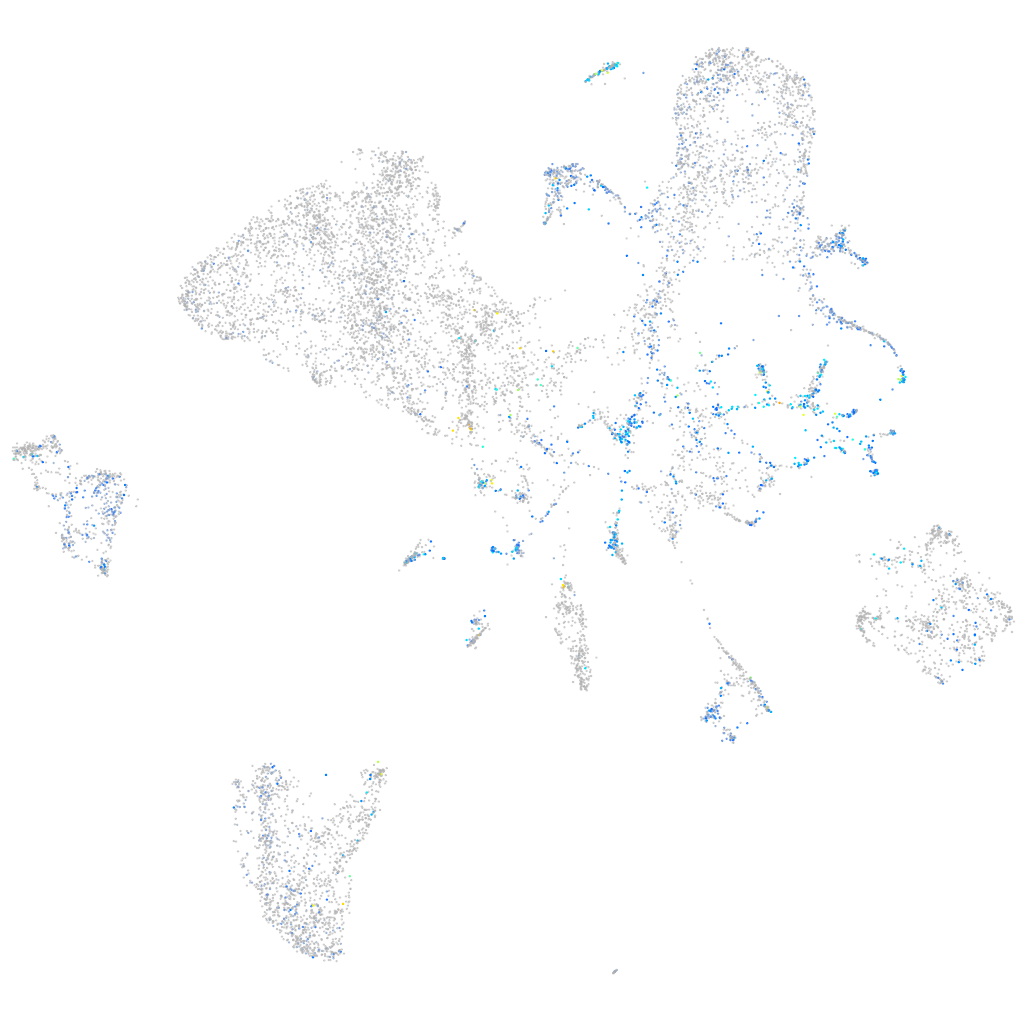
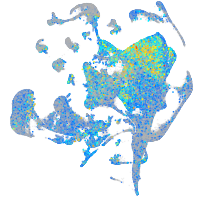
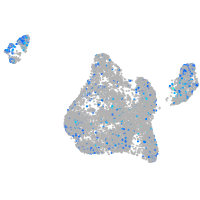
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.310 | gamt | -0.259 |
| ier2b | 0.281 | gatm | -0.242 |
| anxa4 | 0.277 | gapdh | -0.238 |
| neurod1 | 0.270 | bhmt | -0.235 |
| elf3 | 0.264 | mat1a | -0.220 |
| si:ch1073-429i10.3.1 | 0.262 | apoa1b | -0.218 |
| ubc | 0.262 | ahcy | -0.216 |
| insm1a | 0.259 | afp4 | -0.214 |
| sat1a.2 | 0.258 | apoa4b.1 | -0.213 |
| calm1b | 0.254 | fbp1b | -0.212 |
| CR383676.1 | 0.252 | agxtb | -0.208 |
| elovl1a | 0.251 | apoc2 | -0.207 |
| vat1 | 0.251 | apoc1 | -0.204 |
| mir375-2 | 0.251 | scp2a | -0.201 |
| capns1b | 0.249 | serpina1 | -0.198 |
| elovl1b | 0.248 | apoa2 | -0.197 |
| atf4b | 0.247 | rbp2b | -0.197 |
| dusp2 | 0.246 | aqp12 | -0.196 |
| btg1 | 0.245 | fabp10a | -0.195 |
| id4 | 0.245 | serpina1l | -0.195 |
| gapdhs | 0.244 | hao1 | -0.194 |
| jpt1b | 0.243 | tfa | -0.193 |
| lysmd2 | 0.242 | fgb | -0.193 |
| btg2 | 0.241 | zgc:123103 | -0.193 |
| jun | 0.239 | apom | -0.193 |
| oaz1b | 0.239 | hpda | -0.193 |
| mcl1a | 0.236 | pnp4b | -0.192 |
| hepacam2 | 0.235 | LOC110437731 | -0.192 |
| cldnh | 0.234 | ttc36 | -0.192 |
| h3f3c | 0.233 | ambp | -0.191 |
| mir7a-1 | 0.232 | mgst1.2 | -0.190 |
| pax6b | 0.231 | agxta | -0.190 |
| gnb1a | 0.230 | fgg | -0.190 |
| epcam | 0.230 | slc27a2a | -0.189 |
| si:dkey-42i9.4 | 0.227 | igfbp2a | -0.189 |