tRNA phosphotransferase 1
ZFIN 
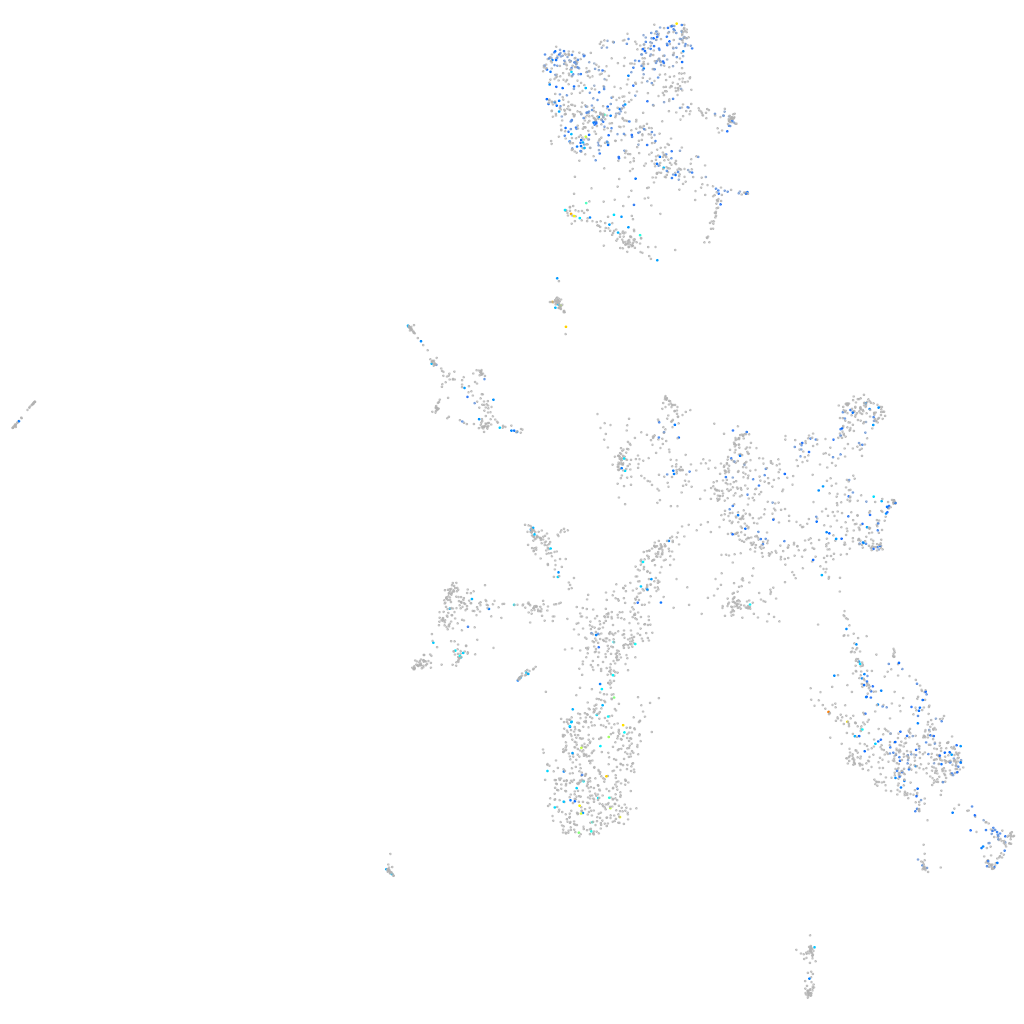
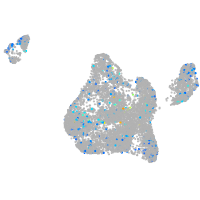
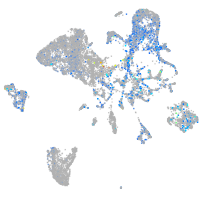
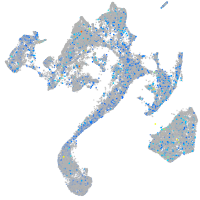
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:66484 | 0.143 | elavl3 | -0.081 |
| CABZ01072157.1 | 0.141 | cxcr4b | -0.069 |
| igfbp5a | 0.136 | CU467822.1 | -0.068 |
| cxcl14 | 0.135 | ebf2 | -0.068 |
| chia.3 | 0.134 | neurod1 | -0.065 |
| anxa11b | 0.130 | FO082781.1 | -0.065 |
| CABZ01039859.1 | 0.130 | ebf3a | -0.064 |
| ahnak | 0.126 | nhlh2 | -0.063 |
| itpkb | 0.126 | sox11a | -0.063 |
| mvp | 0.126 | celf3a | -0.062 |
| CABZ01102518.1 | 0.125 | si:dkey-56m19.5 | -0.062 |
| gna14a | 0.124 | nova2 | -0.060 |
| ponzr6 | 0.123 | sox11b | -0.059 |
| myo6a | 0.122 | myt1a | -0.057 |
| hopx | 0.122 | insm1a | -0.056 |
| calm1b | 0.121 | inka1a | -0.056 |
| scinlb | 0.120 | si:dkey-280e21.3 | -0.056 |
| zgc:154054 | 0.120 | si:ch211-222l21.1 | -0.055 |
| FP236327.1 | 0.120 | si:ch211-193l2.6 | -0.055 |
| rgs1 | 0.119 | NC-002333.4 | -0.055 |
| pax9 | 0.119 | lhx2a | -0.054 |
| fam57ba | 0.118 | ca16b | -0.054 |
| avil | 0.117 | neurod4 | -0.052 |
| s100u | 0.117 | tnni2a.4 | -0.051 |
| rassf6 | 0.117 | tpma | -0.049 |
| gng13a | 0.117 | cdk5r2a | -0.049 |
| vill | 0.117 | RSPH1 | -0.049 |
| myl9a | 0.116 | dla | -0.049 |
| si:ch1073-443f11.2 | 0.116 | dpysl2b | -0.049 |
| gpa33a | 0.115 | tfap2e | -0.048 |
| trpm5 | 0.114 | hnrnpa0a | -0.048 |
| cav1 | 0.114 | her4.4 | -0.048 |
| anxa4 | 0.113 | jakmip2 | -0.048 |
| plecb | 0.113 | CABZ01086877.1 | -0.048 |
| BX601648.2 | 0.113 | add2 | -0.048 |