tripartite motif containing 35-9
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous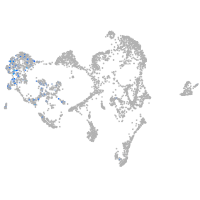 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108183320 | 0.268 | ptmab | -0.085 |
| LOC108190613 | 0.266 | pmp22a | -0.076 |
| cldne | 0.253 | fkbp14 | -0.075 |
| znf185 | 0.251 | c1qtnf5 | -0.073 |
| hrc | 0.249 | mfap2 | -0.071 |
| si:ch211-272b8.6 | 0.248 | fbln1 | -0.067 |
| capn9 | 0.248 | fstl1b | -0.067 |
| lye | 0.243 | tspan7 | -0.066 |
| stard14 | 0.238 | hgd | -0.065 |
| si:ch211-157c3.4 | 0.237 | bhmt | -0.065 |
| si:ch211-195b11.3 | 0.236 | calr | -0.065 |
| si:ch73-204p21.2 | 0.235 | marcksl1a | -0.063 |
| si:ch211-125o16.4 | 0.234 | si:ch211-222l21.1 | -0.063 |
| anxa1c | 0.234 | fthl27 | -0.061 |
| elovl7a | 0.234 | crabp2a | -0.061 |
| osbpl7 | 0.231 | msna | -0.061 |
| zgc:193505 | 0.228 | idh2 | -0.060 |
| icn2 | 0.225 | zgc:153675 | -0.060 |
| si:dkey-87o1.2 | 0.223 | twist1a | -0.060 |
| cldnb | 0.222 | timp2a | -0.059 |
| dhrs13a.2 | 0.220 | hmgb3a | -0.058 |
| abcb5 | 0.220 | prrx1b | -0.058 |
| mgst1.2 | 0.220 | crtap | -0.058 |
| agr1 | 0.219 | ssr3 | -0.058 |
| ppl | 0.219 | tpm4a | -0.058 |
| selenow2b | 0.218 | pcbd1 | -0.058 |
| hsd17b12a | 0.218 | fkbp7 | -0.058 |
| scel | 0.217 | ntd5 | -0.057 |
| zgc:111983 | 0.217 | fkbp10b | -0.057 |
| anxa1b | 0.216 | ptx3a | -0.057 |
| cx30.3 | 0.216 | ttc36 | -0.057 |
| si:ch73-52f15.5 | 0.215 | prrx1a | -0.057 |
| evpla | 0.215 | cirbpb | -0.057 |
| si:dkey-175d9.2 | 0.215 | zgc:66479 | -0.056 |
| zgc:110333 | 0.215 | nucb2a | -0.056 |