tripartite motif containing 35-7
ZFIN 
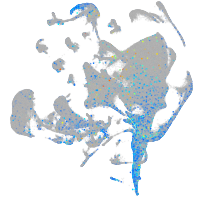
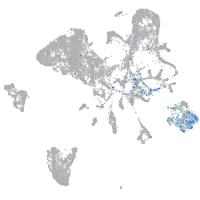
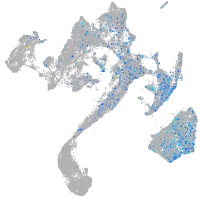
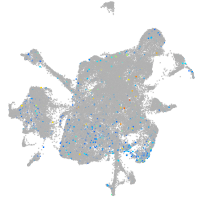
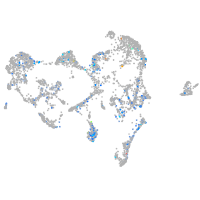
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-205h11.1 | 0.079 | ckbb | -0.044 |
| pcna | 0.073 | myt1b | -0.033 |
| dut | 0.069 | slc1a2b | -0.033 |
| nasp | 0.068 | rtn1a | -0.032 |
| stmn1a | 0.067 | si:dkeyp-75h12.5 | -0.032 |
| zgc:110540 | 0.067 | rnasekb | -0.030 |
| nutf2l | 0.066 | gapdhs | -0.028 |
| fen1 | 0.066 | cotl1 | -0.028 |
| rrm1 | 0.065 | mllt11 | -0.028 |
| chaf1a | 0.063 | aldocb | -0.027 |
| unga | 0.062 | elavl3 | -0.026 |
| si:dkey-41c6.3 | 0.061 | tnrc6c1 | -0.026 |
| rpa2 | 0.061 | efhd1 | -0.026 |
| anp32b | 0.060 | nhlh2 | -0.026 |
| ranbp1 | 0.060 | nova2 | -0.026 |
| FO834903.1 | 0.060 | slc6a1b | -0.026 |
| dnajc9 | 0.060 | syt11a | -0.026 |
| si:ch1073-13h15.3 | 0.059 | pvalb1 | -0.025 |
| chtf8 | 0.059 | gpm6aa | -0.025 |
| rrm2 | 0.059 | anxa13 | -0.025 |
| mcm7 | 0.059 | slc40a1 | -0.025 |
| lig1 | 0.058 | hbae1.1 | -0.025 |
| slbp | 0.058 | pvalb2 | -0.024 |
| cdx4 | 0.058 | ppdpfb | -0.024 |
| rnaseh2a | 0.057 | gabarapl2 | -0.024 |
| mir733 | 0.057 | fabp7a | -0.024 |
| dek | 0.057 | slc4a4a | -0.024 |
| shmt1 | 0.057 | vamp2 | -0.024 |
| cdc6 | 0.057 | acbd7 | -0.024 |
| adssl | 0.056 | cadm4 | -0.023 |
| atad2 | 0.056 | tspan7b | -0.023 |
| znfl2a | 0.056 | gpm6ab | -0.023 |
| gins3 | 0.055 | ndrg3a | -0.023 |
| mis12 | 0.055 | rufy3 | -0.023 |
| rfc4 | 0.054 | CU467822.1 | -0.023 |