tribbles pseudokinase 3
ZFIN 
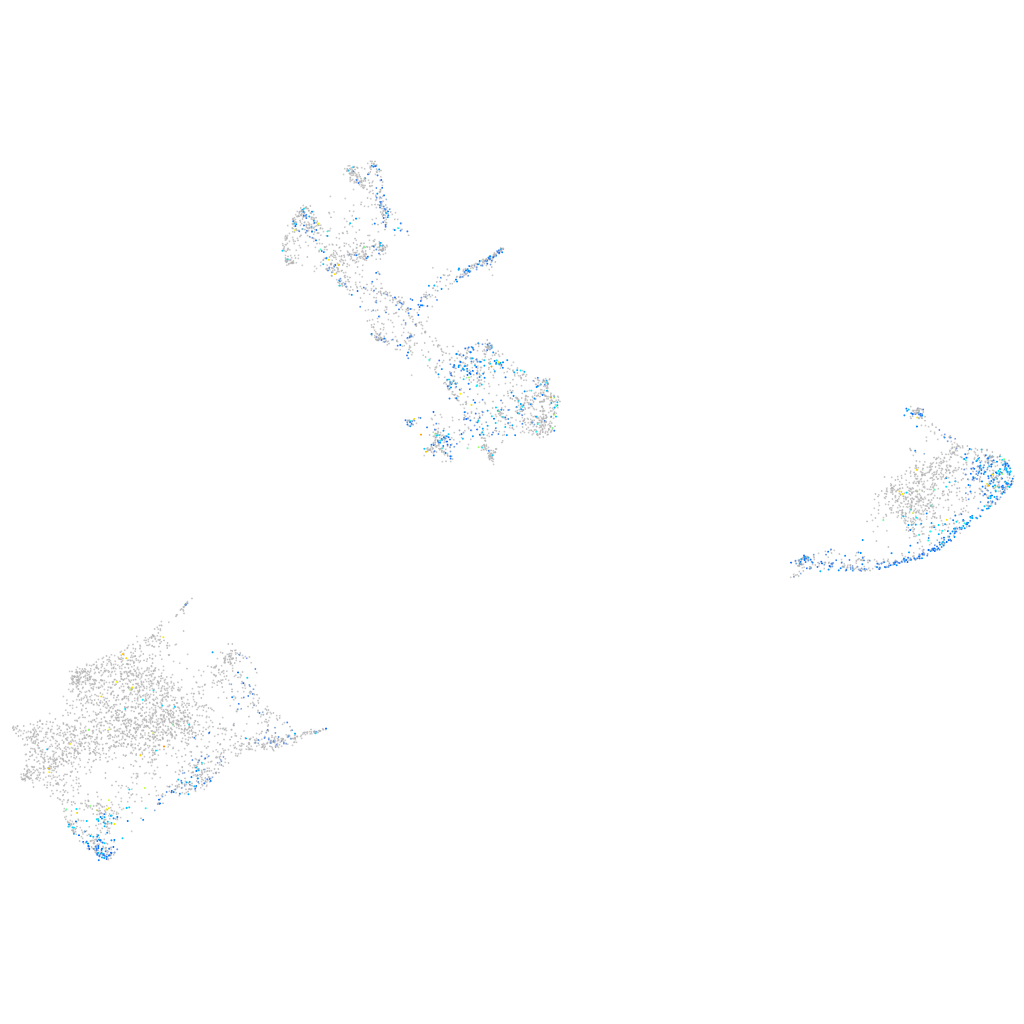
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.227 | CABZ01021592.1 | -0.172 |
| CR383676.1 | 0.226 | si:dkey-251i10.2 | -0.159 |
| kita | 0.192 | paics | -0.155 |
| CR847566.1 | 0.185 | uraha | -0.150 |
| ddit3 | 0.182 | mdh1aa | -0.140 |
| si:ch211-195b13.1 | 0.180 | prdx1 | -0.121 |
| slc7a5 | 0.179 | cyb5a | -0.120 |
| ier5 | 0.177 | impdh1b | -0.117 |
| mcl1a | 0.177 | gch2 | -0.111 |
| dusp1 | 0.176 | tmem130 | -0.110 |
| gadd45ga | 0.173 | akr1b1 | -0.110 |
| atf4b | 0.171 | gstp1 | -0.110 |
| aadac | 0.169 | pts | -0.109 |
| gadd45ba | 0.169 | atic | -0.107 |
| socs3a | 0.166 | rbp4l | -0.106 |
| oca2 | 0.164 | gpd1b | -0.100 |
| sdc4 | 0.164 | slc22a7a | -0.098 |
| ccng2 | 0.163 | hbbe1.3 | -0.094 |
| slc6a15 | 0.161 | sult1st1 | -0.093 |
| slc25a55a | 0.160 | bco1 | -0.091 |
| dct | 0.160 | aox5 | -0.091 |
| tyrp1a | 0.160 | prdx5 | -0.089 |
| slc3a2a | 0.160 | hbae3 | -0.087 |
| slc24a5 | 0.159 | glulb | -0.083 |
| si:zfos-943e10.1 | 0.159 | hbae1.1 | -0.082 |
| ier2a | 0.159 | zgc:153031 | -0.079 |
| btg2 | 0.158 | prps1a | -0.077 |
| ubb | 0.158 | aldob | -0.077 |
| tfap2e | 0.158 | phyhd1 | -0.077 |
| tyrp1b | 0.158 | pvalb2 | -0.075 |
| si:ch73-389b16.1 | 0.157 | sprb | -0.074 |
| mchr2 | 0.157 | oacyl | -0.072 |
| lamp1a | 0.157 | actc1b | -0.072 |
| fosab | 0.154 | zgc:110339 | -0.069 |
| klf6a | 0.153 | slc2a15a | -0.068 |