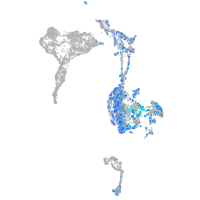
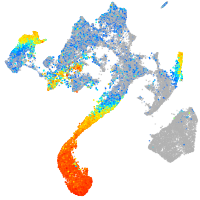
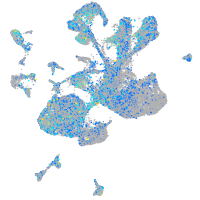
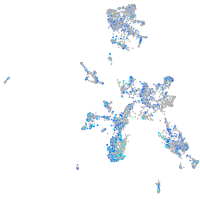
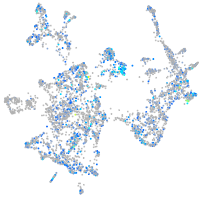
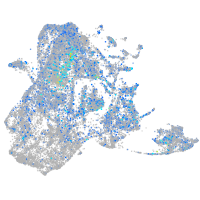
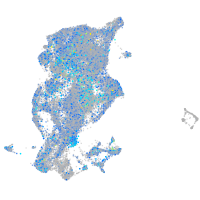
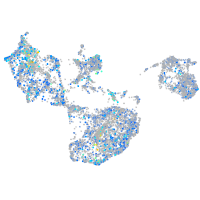
alpha-tropomyosin
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pvalb2 | 0.302 | rps12 | -0.118 |
| pvalb1 | 0.287 | rps18 | -0.107 |
| actc1b | 0.285 | uba52 | -0.104 |
| mylpfa | 0.262 | rpl23a | -0.104 |
| mylz3 | 0.232 | cfl1 | -0.100 |
| tnni2a.4 | 0.208 | pabpc1a | -0.091 |
| tnnt3b | 0.189 | rps8a | -0.090 |
| mylpfb | 0.173 | eif2s2 | -0.089 |
| apoa2 | 0.166 | mibp2 | -0.089 |
| krt4 | 0.165 | rpl32 | -0.088 |
| myhz1.1 | 0.165 | rpl22l1 | -0.086 |
| ckma | 0.160 | seta | -0.086 |
| ckmb | 0.156 | rplp2l | -0.085 |
| nme2b.2 | 0.154 | hmga1a | -0.085 |
| cyt1 | 0.147 | rpl18a | -0.084 |
| apoa1b | 0.144 | snrpb | -0.084 |
| myl1 | 0.143 | si:ch211-222l21.1 | -0.083 |
| acta1b | 0.141 | rpl36 | -0.082 |
| cyt1l | 0.122 | rpl17 | -0.082 |
| myhz2 | 0.122 | snrpd1 | -0.082 |
| prss59.2 | 0.122 | zgc:153409 | -0.080 |
| prss1 | 0.118 | sub1a | -0.080 |
| ttn.1 | 0.118 | snrpg | -0.079 |
| tnnc2 | 0.113 | rpl3 | -0.079 |
| CR383676.1 | 0.110 | rps3 | -0.078 |
| si:ch211-195b11.3 | 0.110 | rps6 | -0.078 |
| krtt1c19e | 0.110 | eif5a2 | -0.077 |
| icn2 | 0.109 | rpl15 | -0.076 |
| ttn.2 | 0.108 | ptges3b | -0.076 |
| myhz1.2 | 0.107 | zgc:114188 | -0.076 |
| si:ch73-367p23.2 | 0.104 | cirbpa | -0.076 |
| gapdh | 0.102 | rpl31 | -0.075 |
| icn | 0.101 | rps11 | -0.075 |
| myl13 | 0.100 | h2afvb | -0.074 |
| ak1 | 0.099 | rpl27 | -0.073 |