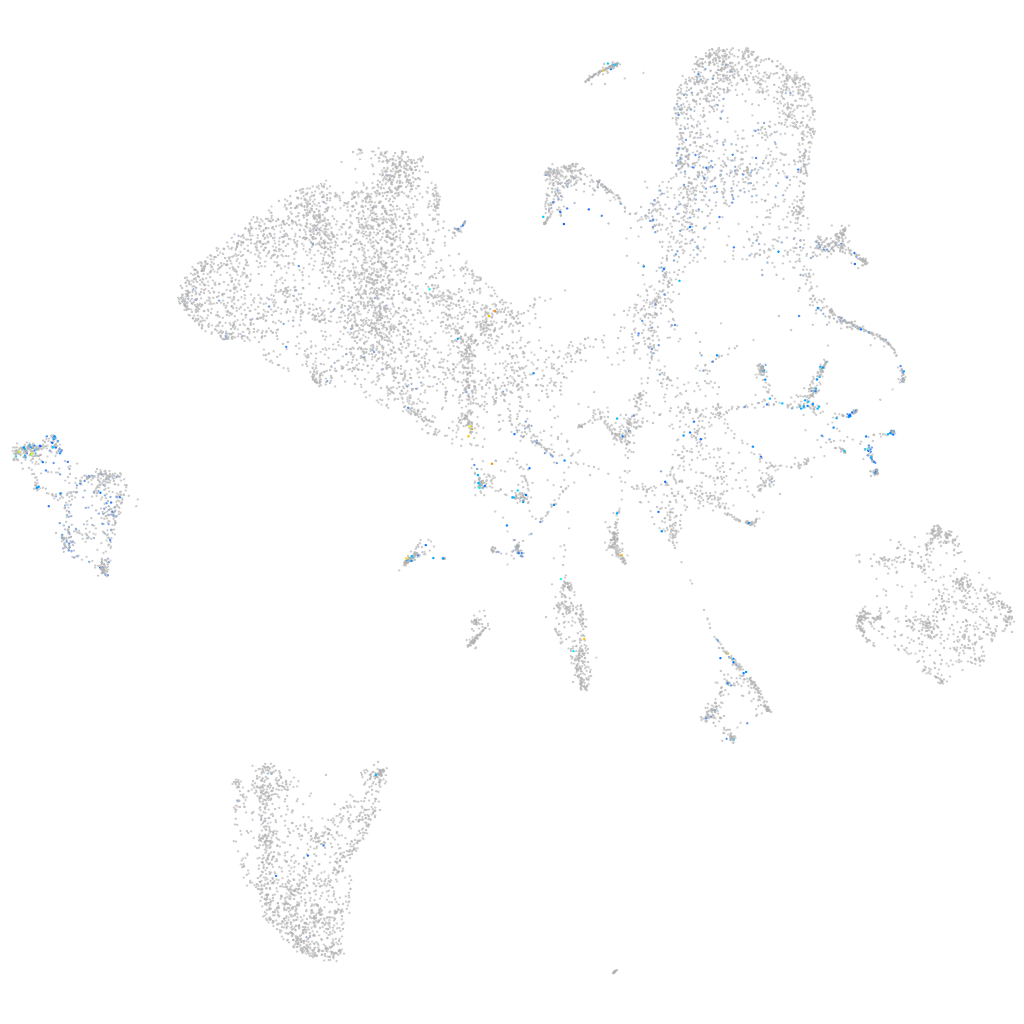
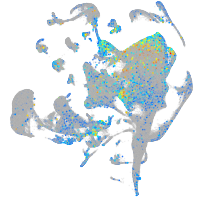
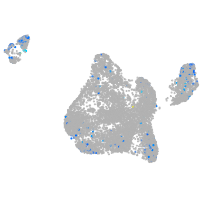
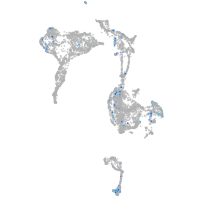
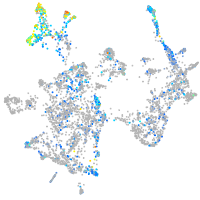
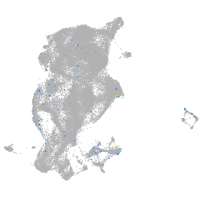
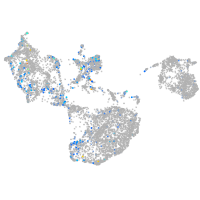
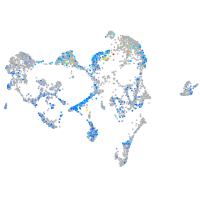
triosephosphate isomerase 1a
ZFIN 
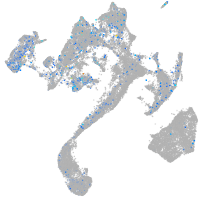
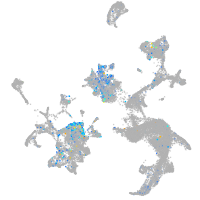
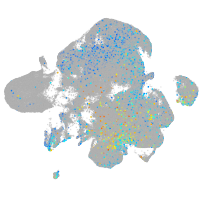
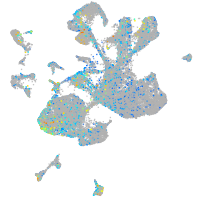
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.197 | gatm | -0.133 |
| rnasekb | 0.183 | gamt | -0.132 |
| calm1b | 0.181 | bhmt | -0.121 |
| calm1a | 0.176 | gapdh | -0.120 |
| scg3 | 0.172 | aqp12 | -0.120 |
| tmed4 | 0.170 | apoc1 | -0.116 |
| eno1a | 0.161 | mat1a | -0.115 |
| scgn | 0.157 | agxtb | -0.111 |
| vat1 | 0.156 | pnp4b | -0.106 |
| icn | 0.154 | ttc36 | -0.105 |
| vamp2 | 0.154 | abat | -0.105 |
| gmds | 0.154 | ahcy | -0.104 |
| ier2b | 0.154 | fbp1b | -0.103 |
| fev | 0.153 | cpn1 | -0.101 |
| tmed7 | 0.152 | ambp | -0.101 |
| CR392036.1 | 0.151 | serpina1l | -0.099 |
| neurod1 | 0.150 | g6pca.2 | -0.099 |
| rprmb | 0.150 | apoa1b | -0.098 |
| cpe | 0.150 | ttr | -0.098 |
| gnb1a | 0.150 | agxta | -0.098 |
| gpc1a | 0.149 | grhprb | -0.098 |
| si:ch1073-429i10.3.1 | 0.149 | fetub | -0.097 |
| mir375-2 | 0.149 | kng1 | -0.097 |
| pax6b | 0.146 | gc | -0.097 |
| chga | 0.146 | zgc:123103 | -0.096 |
| c2cd4a | 0.145 | serpina1 | -0.096 |
| aldoca | 0.145 | apoa4b.1 | -0.096 |
| olfm2a | 0.144 | LOC110437731 | -0.096 |
| LOC101882249 | 0.143 | ugp2a | -0.095 |
| syt1a | 0.143 | rbp2b | -0.095 |
| cd63 | 0.143 | hpda | -0.094 |
| si:ch211-165b10.3 | 0.142 | f2 | -0.094 |
| lin7b | 0.141 | ces2 | -0.094 |
| atpv0e2 | 0.141 | fgg | -0.093 |
| cnih3 | 0.141 | cfhl4 | -0.093 |