thymocyte selection-associated high mobility group box
ZFIN 
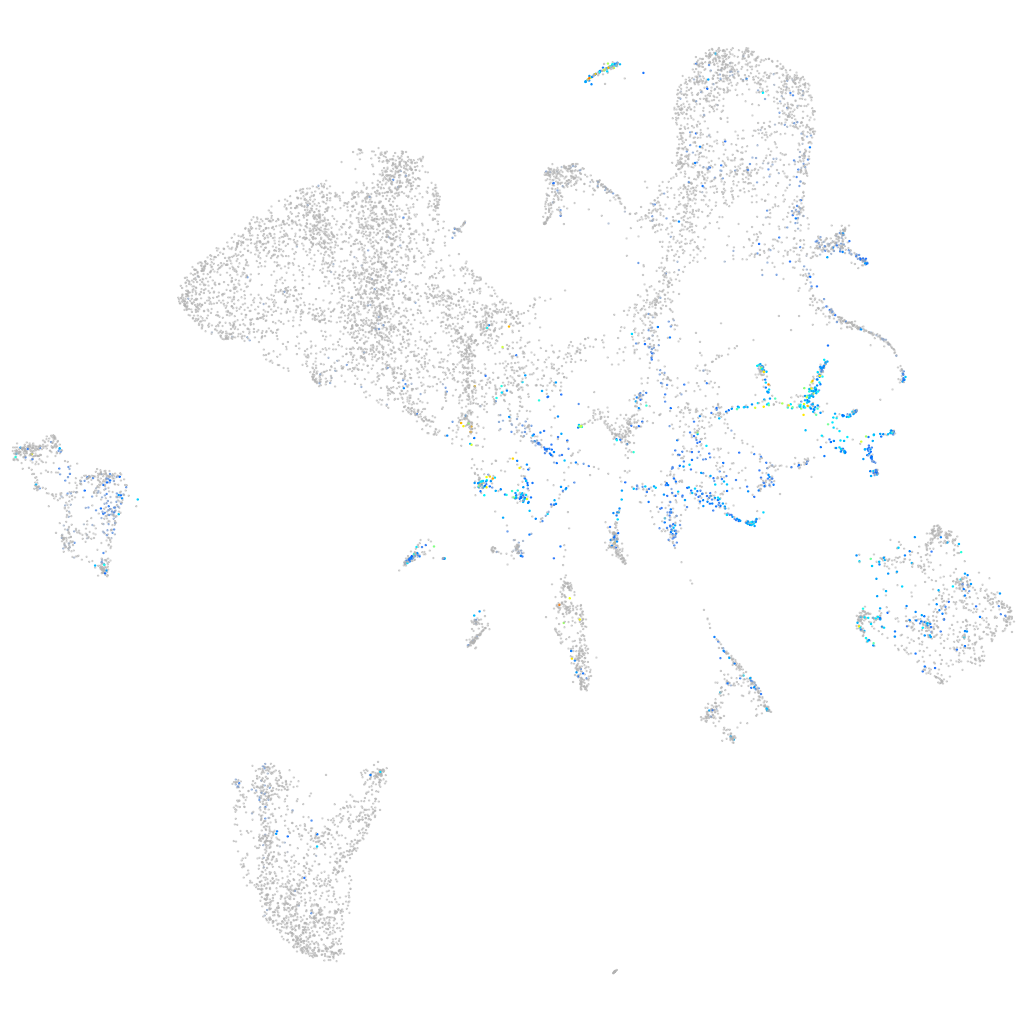
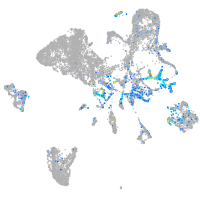
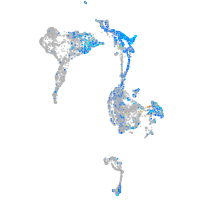
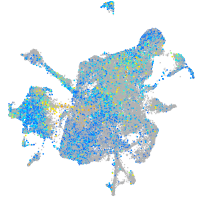
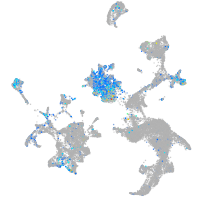
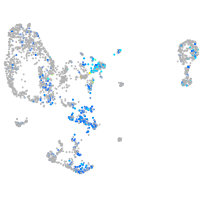
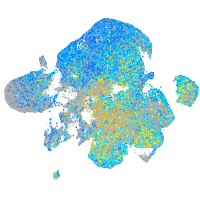
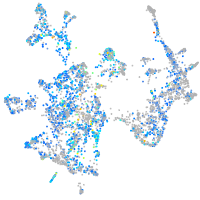
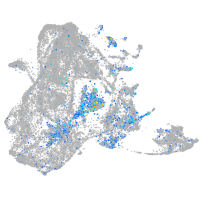
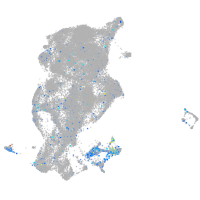
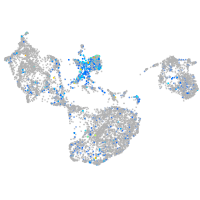
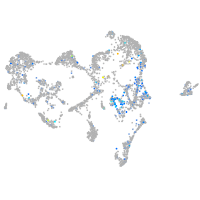
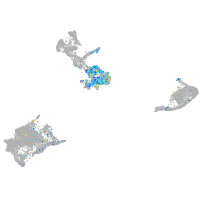
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.435 | gapdh | -0.284 |
| neurod1 | 0.403 | ahcy | -0.273 |
| insm1a | 0.362 | gamt | -0.272 |
| pax6b | 0.356 | gatm | -0.240 |
| c2cd4a | 0.356 | mat1a | -0.233 |
| pcsk1nl | 0.346 | fbp1b | -0.232 |
| fev | 0.345 | eno3 | -0.227 |
| egr4 | 0.343 | aldob | -0.227 |
| myt1b | 0.341 | cx32.3 | -0.227 |
| mir375-2 | 0.338 | apoa4b.1 | -0.224 |
| mir7a-1 | 0.338 | glud1b | -0.222 |
| nkx2.2a | 0.337 | gstt1a | -0.217 |
| lysmd2 | 0.335 | scp2a | -0.214 |
| scgn | 0.330 | apoc2 | -0.214 |
| syt1a | 0.319 | nupr1b | -0.212 |
| vamp2 | 0.319 | aldh6a1 | -0.212 |
| dll4 | 0.318 | agxtb | -0.212 |
| jpt1b | 0.314 | apoa1b | -0.211 |
| arxa | 0.310 | aldh7a1 | -0.205 |
| hepacam2 | 0.304 | bhmt | -0.204 |
| hmgn6 | 0.302 | suclg1 | -0.204 |
| isl1 | 0.301 | agxta | -0.204 |
| insm1b | 0.300 | pnp4b | -0.201 |
| vat1 | 0.299 | mgst1.2 | -0.199 |
| si:dkey-42i9.4 | 0.298 | abat | -0.196 |
| id4 | 0.297 | ckba | -0.196 |
| tspan7 | 0.295 | ugt1a7 | -0.194 |
| hmgb3a | 0.295 | nipsnap3a | -0.194 |
| si:dkey-153k10.9 | 0.293 | gcshb | -0.192 |
| tspan7b | 0.290 | gnmt | -0.192 |
| ret | 0.289 | tdo2a | -0.192 |
| LOC100537384 | 0.288 | afp4 | -0.191 |
| elovl1a | 0.287 | hdlbpa | -0.190 |
| rnasekb | 0.283 | fabp10a | -0.189 |
| si:ch1073-429i10.3.1 | 0.282 | pgm1 | -0.188 |