translocase of outer mitochondrial membrane 20b
ZFIN 
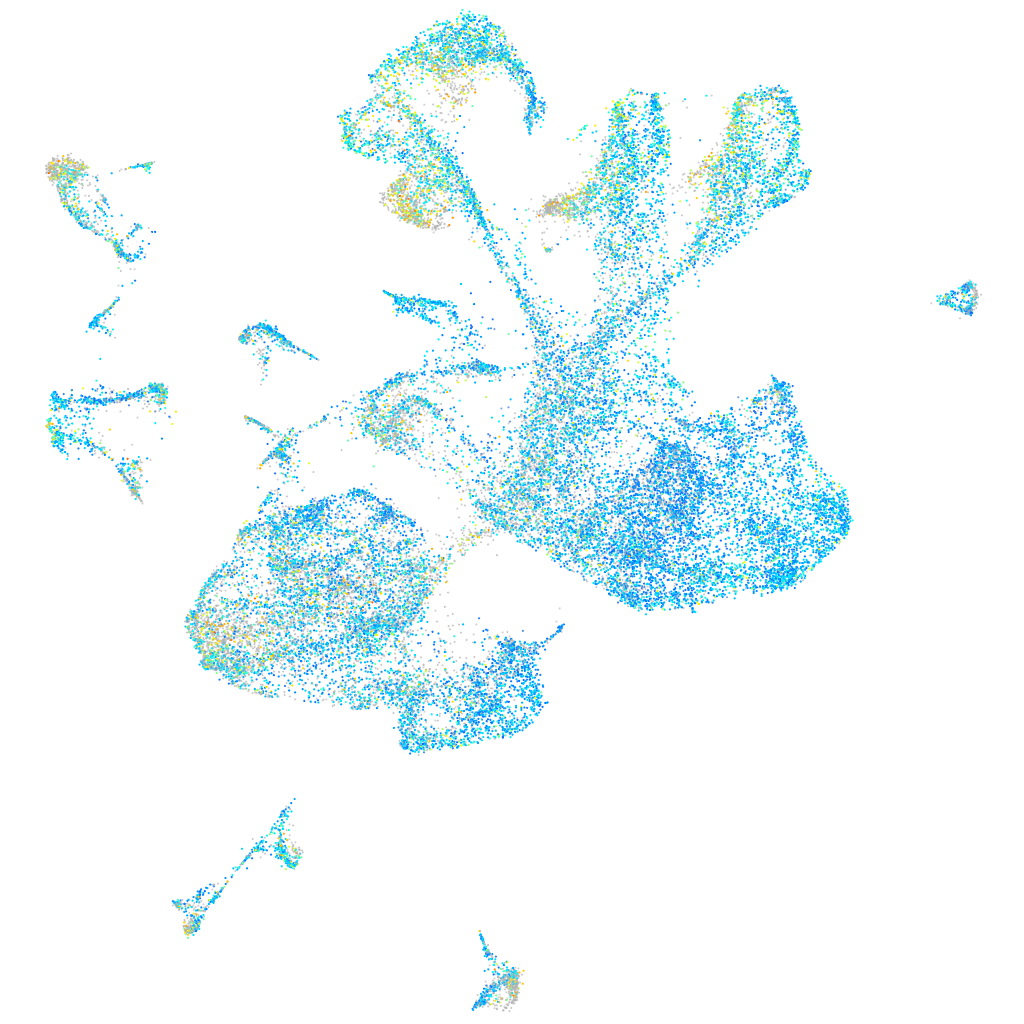
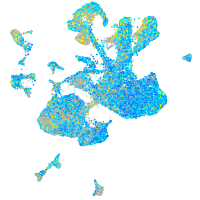
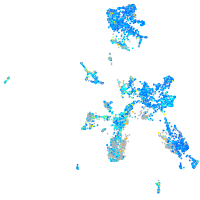
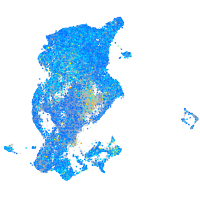
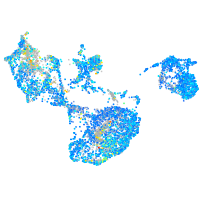
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc25a5 | 0.195 | atp1a1b | -0.154 |
| vdac1 | 0.190 | fabp7a | -0.137 |
| hsp90ab1 | 0.186 | CU467822.1 | -0.135 |
| naca | 0.171 | gpm6bb | -0.133 |
| hnrnpaba | 0.170 | si:ch211-251b21.1 | -0.121 |
| hspa8 | 0.170 | slc1a2b | -0.106 |
| hnrnpa0l | 0.170 | notch3 | -0.104 |
| btf3 | 0.167 | zgc:165461 | -0.103 |
| COX5B | 0.167 | zgc:158463 | -0.103 |
| eif3f | 0.167 | atp1b4 | -0.103 |
| vdac2 | 0.166 | ptn | -0.100 |
| rsl24d1 | 0.165 | qki2 | -0.094 |
| eef1db | 0.164 | slc4a4a | -0.093 |
| slc25a3b | 0.164 | mdka | -0.093 |
| khdrbs1a | 0.163 | slc6a9 | -0.091 |
| h3f3d | 0.162 | gpr37l1b | -0.089 |
| aldoaa | 0.161 | CU634008.1 | -0.089 |
| eif3ba | 0.160 | CR848047.1 | -0.089 |
| calm3b | 0.159 | cx43 | -0.089 |
| cct5 | 0.158 | ppap2d | -0.087 |
| sumo2b | 0.158 | glula | -0.087 |
| cct2 | 0.158 | pvalb1 | -0.084 |
| eef1a1l1 | 0.157 | actc1b | -0.082 |
| cycsb | 0.157 | slc1a3b | -0.082 |
| rpl19 | 0.156 | pvalb2 | -0.081 |
| sumo3a | 0.154 | cldn19 | -0.080 |
| tcp1 | 0.152 | s1pr1 | -0.080 |
| oaz1b | 0.151 | efhd1 | -0.079 |
| cct6a | 0.151 | npas1 | -0.078 |
| cct4 | 0.150 | mdkb | -0.078 |
| cct7 | 0.150 | ptprz1b | -0.076 |
| eef1g | 0.150 | gpm6aa | -0.076 |
| cirbpb | 0.150 | mt2 | -0.076 |
| cox4i1 | 0.149 | si:ch211-66e2.5 | -0.073 |
| ube2e1 | 0.148 | slc6a11b | -0.073 |