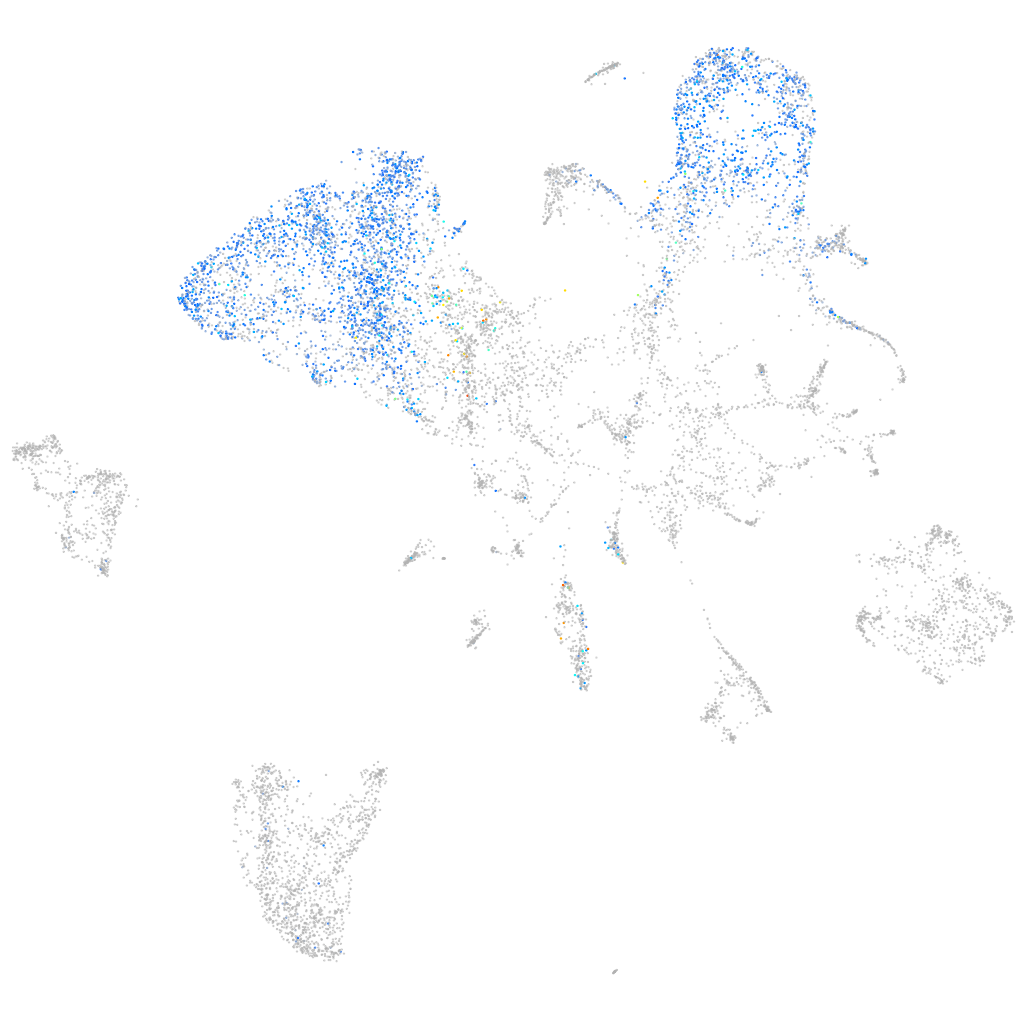
transmembrane protein 82
ZFIN 
Other cell groups


all cells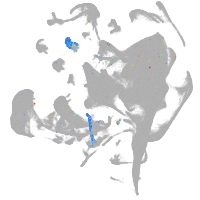 |
blastomeres |
PGCs |
endoderm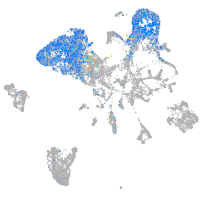 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 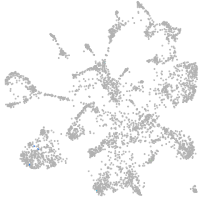 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 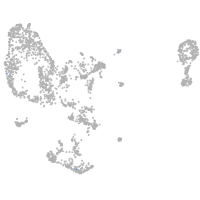 |
neural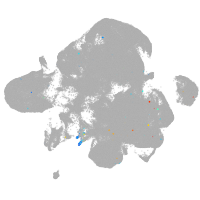 |
spinal cord / glia |
eye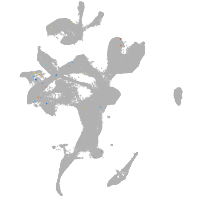 |
taste / olfactory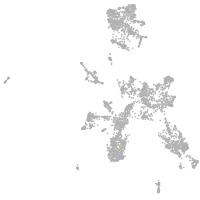 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoa4b.1 | 0.496 | si:ch211-222l21.1 | -0.272 |
| gpx4a | 0.481 | hmgn6 | -0.253 |
| scp2a | 0.472 | tuba8l4 | -0.248 |
| rdh1 | 0.471 | epcam | -0.243 |
| dhrs9 | 0.470 | slc38a5b | -0.243 |
| apoc2 | 0.468 | h3f3d | -0.241 |
| apobb.1 | 0.451 | idh2 | -0.240 |
| glud1b | 0.449 | si:ch73-1a9.3 | -0.239 |
| ugt5b4 | 0.447 | hmgb3a | -0.237 |
| srd5a2a | 0.446 | hmgn2 | -0.229 |
| cyp8b1 | 0.445 | h2afvb | -0.228 |
| slc37a4a | 0.445 | hmgb2a | -0.228 |
| mgst1.2 | 0.441 | ywhaz | -0.228 |
| gapdh | 0.440 | cirbpa | -0.227 |
| suclg1 | 0.437 | syncrip | -0.223 |
| slco1d1 | 0.436 | rtn1a | -0.221 |
| afp4 | 0.434 | khdrbs1a | -0.220 |
| gcshb | 0.428 | hnrnpaba | -0.219 |
| sult2st2 | 0.428 | ptmab | -0.216 |
| sdr16c5b | 0.427 | glulb | -0.214 |
| cyp4v8 | 0.426 | cirbpb | -0.213 |
| ugt1a7 | 0.424 | jpt1b | -0.212 |
| g6pca.2 | 0.422 | nucks1a | -0.209 |
| gstt1a | 0.421 | hmgb1b | -0.208 |
| msrb2 | 0.420 | si:ch73-281n10.2 | -0.207 |
| eno3 | 0.420 | hmgb2b | -0.205 |
| fbp1b | 0.419 | marcksl1b | -0.205 |
| mdh1aa | 0.418 | ubc | -0.205 |
| haao | 0.416 | cldn7b | -0.204 |
| pla2g12b | 0.414 | cldnb | -0.203 |
| apoa1b | 0.413 | si:ch211-195b11.3 | -0.202 |
| etnppl | 0.412 | seta | -0.202 |
| tdo2a | 0.411 | stmn1a | -0.200 |
| sod1 | 0.410 | hmgb1a | -0.200 |
| tpi1b | 0.406 | marcksb | -0.198 |