transmembrane protein 65
ZFIN 
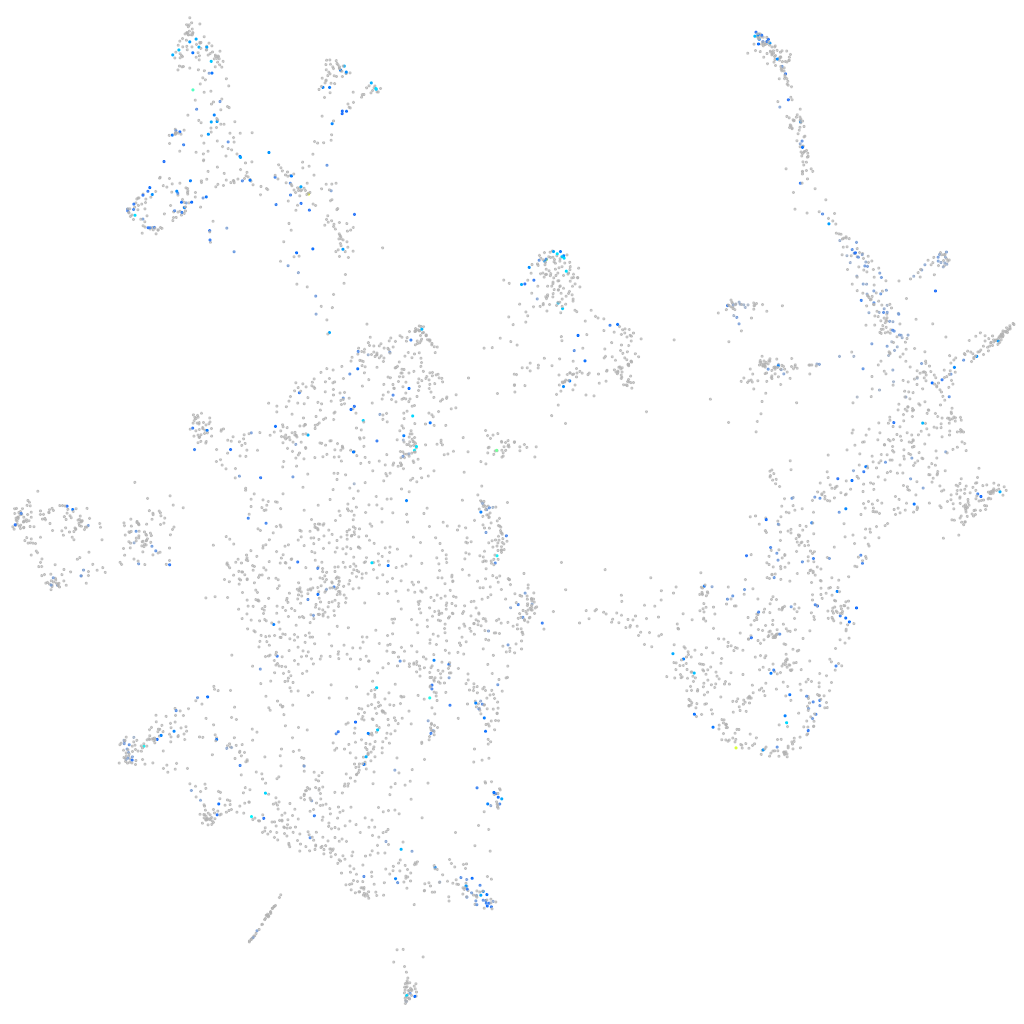
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.159 | si:dkey-151g10.6 | -0.151 |
| si:ch73-15n15.3 | 0.158 | rplp1 | -0.149 |
| aldocb | 0.156 | rplp2l | -0.148 |
| vamp2 | 0.156 | rpl37 | -0.140 |
| rnasekb | 0.155 | rps28 | -0.136 |
| ndufa4 | 0.155 | rpl36a | -0.135 |
| gapdhs | 0.153 | rps20 | -0.132 |
| nptna | 0.148 | rpl39 | -0.129 |
| cpeb4b | 0.146 | rpl23 | -0.128 |
| calm1a | 0.145 | rpl28 | -0.126 |
| osbpl1a | 0.145 | rps9 | -0.123 |
| gfi1aa | 0.144 | rps15a | -0.123 |
| spa17 | 0.144 | rpl7a | -0.121 |
| laptm4b | 0.143 | rps29 | -0.121 |
| atp2b1a | 0.143 | rps24 | -0.120 |
| zgc:194578 | 0.143 | rps21 | -0.119 |
| XLOC-025107 | 0.142 | rps23 | -0.118 |
| myo6b | 0.141 | rpl17 | -0.117 |
| gdi1 | 0.141 | rpl35a | -0.117 |
| calm1b | 0.139 | rpl29 | -0.116 |
| ckbb | 0.138 | rps2 | -0.115 |
| LO018422.1 | 0.138 | rps10 | -0.115 |
| tmc2a | 0.137 | rpl36 | -0.115 |
| pygmb | 0.136 | rps19 | -0.114 |
| otofb | 0.135 | rpl26 | -0.114 |
| pcyt2 | 0.135 | rps12 | -0.111 |
| calml4a | 0.135 | rpl7 | -0.111 |
| apba1a | 0.134 | rps14 | -0.110 |
| eps8b | 0.134 | rpl18 | -0.109 |
| CR925719.1 | 0.134 | rpl38 | -0.109 |
| uqcrfs1 | 0.134 | rps27.1 | -0.108 |
| si:ch211-236p22.1 | 0.133 | rps26 | -0.107 |
| grxcr2 | 0.133 | rpl21 | -0.107 |
| atp6v1g1 | 0.133 | rpl10a | -0.106 |
| atp1a3b | 0.133 | rpl9 | -0.106 |