transmembrane protein 63C
ZFIN 
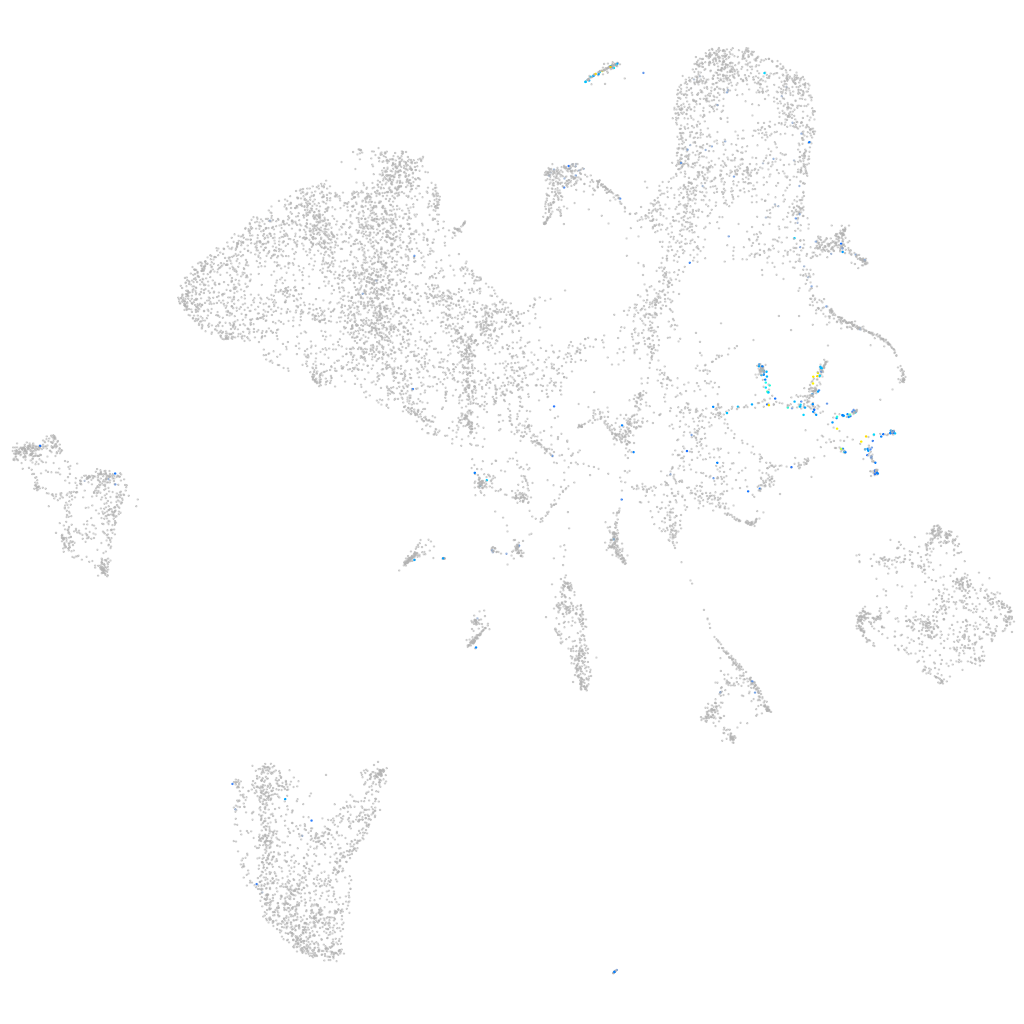
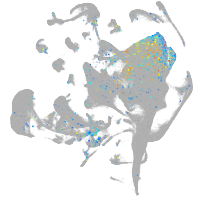
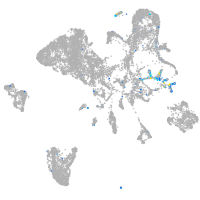
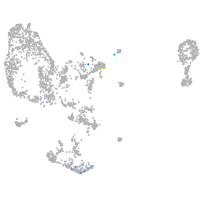
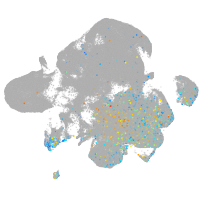
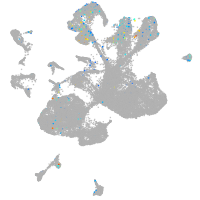
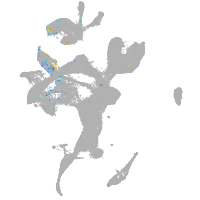
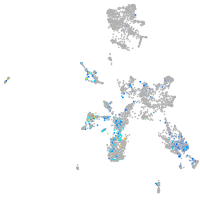
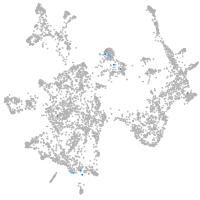
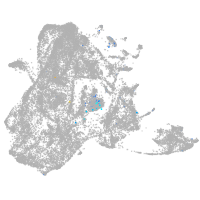
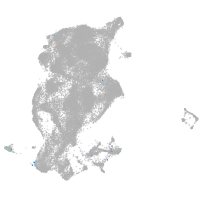
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.394 | ahcy | -0.120 |
| neurod1 | 0.366 | gapdh | -0.117 |
| pax6b | 0.364 | gamt | -0.107 |
| mir7a-1 | 0.344 | gatm | -0.103 |
| pcsk1nl | 0.333 | hdlbpa | -0.099 |
| scgn | 0.331 | bhmt | -0.098 |
| gpc1a | 0.315 | aldob | -0.096 |
| zgc:101731 | 0.309 | si:dkey-16p21.8 | -0.095 |
| fev | 0.306 | apoa1b | -0.095 |
| rprmb | 0.306 | gstr | -0.094 |
| tspan7b | 0.303 | fbp1b | -0.093 |
| isl1 | 0.303 | fabp3 | -0.093 |
| cacna1c | 0.302 | mat1a | -0.093 |
| egr4 | 0.301 | gstt1a | -0.092 |
| id4 | 0.300 | aldh7a1 | -0.092 |
| si:dkey-153k10.9 | 0.294 | apoc2 | -0.091 |
| scg2a | 0.293 | aldh6a1 | -0.091 |
| hepacam2 | 0.290 | nupr1b | -0.090 |
| vat1 | 0.288 | apoa4b.1 | -0.089 |
| pygma | 0.288 | apoc1 | -0.088 |
| c2cd4a | 0.284 | shmt1 | -0.086 |
| syt1a | 0.284 | aqp12 | -0.086 |
| rasd4 | 0.284 | cx32.3 | -0.085 |
| cplx2l | 0.279 | afp4 | -0.084 |
| pcsk1 | 0.279 | hspe1 | -0.084 |
| kcnj11 | 0.278 | apoa2 | -0.084 |
| insm1a | 0.278 | scp2a | -0.084 |
| mir375-2 | 0.275 | agxtb | -0.083 |
| lysmd2 | 0.272 | tfa | -0.083 |
| syt11a | 0.265 | agxta | -0.083 |
| vamp2 | 0.263 | gnmt | -0.083 |
| gnao1a | 0.257 | ppa1b | -0.082 |
| rnasekb | 0.256 | gstp1 | -0.081 |
| camk1da | 0.253 | aldh1l1 | -0.080 |
| foxo1a | 0.249 | mgst1.2 | -0.080 |