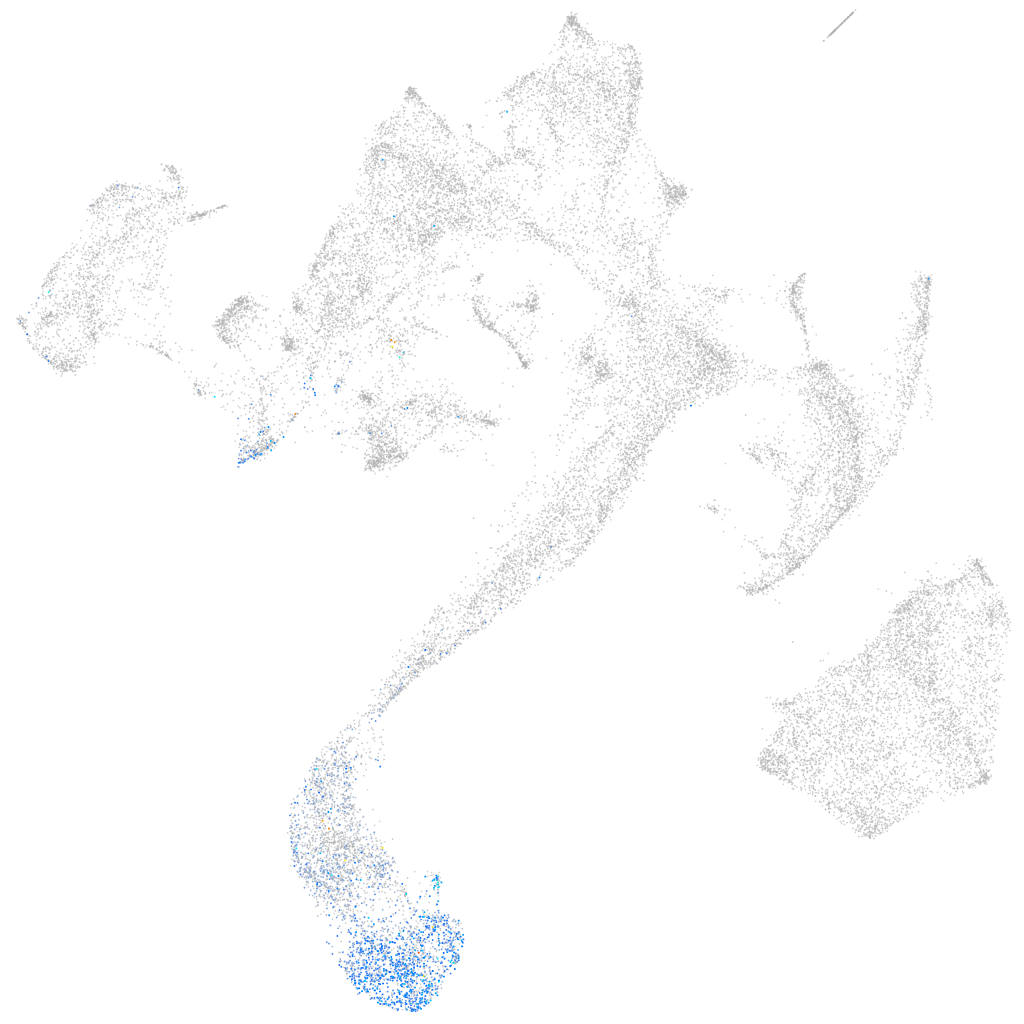
transmembrane protein 41ab
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ampd1 | 0.579 | h3f3d | -0.407 |
| slc25a4 | 0.567 | cirbpb | -0.388 |
| igfn1.3 | 0.541 | hsp90ab1 | -0.377 |
| atp2a1l | 0.536 | naca | -0.366 |
| CABZ01061524.1 | 0.529 | pabpc1a | -0.360 |
| pvalb1 | 0.525 | h2afvb | -0.351 |
| pvalb2 | 0.525 | khdrbs1a | -0.350 |
| hsc70 | 0.517 | hmgn6 | -0.343 |
| tnnt3b | 0.517 | hnrnpaba | -0.337 |
| mylpfb | 0.514 | si:ch73-1a9.3 | -0.336 |
| casq1a | 0.507 | actb2 | -0.334 |
| eef2l2 | 0.507 | ran | -0.332 |
| tnni2a.4 | 0.501 | hmga1a | -0.332 |
| si:ch211-255p10.3 | 0.501 | eef1a1l1 | -0.328 |
| myoz1b | 0.495 | fthl27 | -0.323 |
| myom2a | 0.494 | cirbpa | -0.321 |
| mylz3 | 0.492 | cfl1 | -0.316 |
| si:ch73-367p23.2 | 0.476 | hmgn2 | -0.313 |
| ryr3 | 0.465 | ubc | -0.311 |
| hhatla | 0.460 | hnrnpa0b | -0.309 |
| gyg1b | 0.457 | snrpf | -0.306 |
| myoz2a | 0.452 | hmgb2b | -0.306 |
| tpma | 0.451 | snrpb | -0.303 |
| pkmb | 0.448 | hspa8 | -0.298 |
| actn3a | 0.447 | hnrnpabb | -0.298 |
| mylpfa | 0.446 | ptmab | -0.297 |
| ankrd1b | 0.440 | setb | -0.296 |
| tnnc2 | 0.437 | si:ch211-222l21.1 | -0.295 |
| si:ch73-366l1.5 | 0.432 | si:ch73-281n10.2 | -0.293 |
| myoz1a | 0.432 | hmgb2a | -0.292 |
| si:dkey-51e6.1 | 0.431 | eef2b | -0.289 |
| rgcc | 0.424 | rbm8a | -0.288 |
| pfkma | 0.421 | hmgn7 | -0.287 |
| myhz2 | 0.419 | syncrip | -0.286 |
| fgf21 | 0.419 | rpl32 | -0.284 |