transmembrane protein 187
ZFIN 
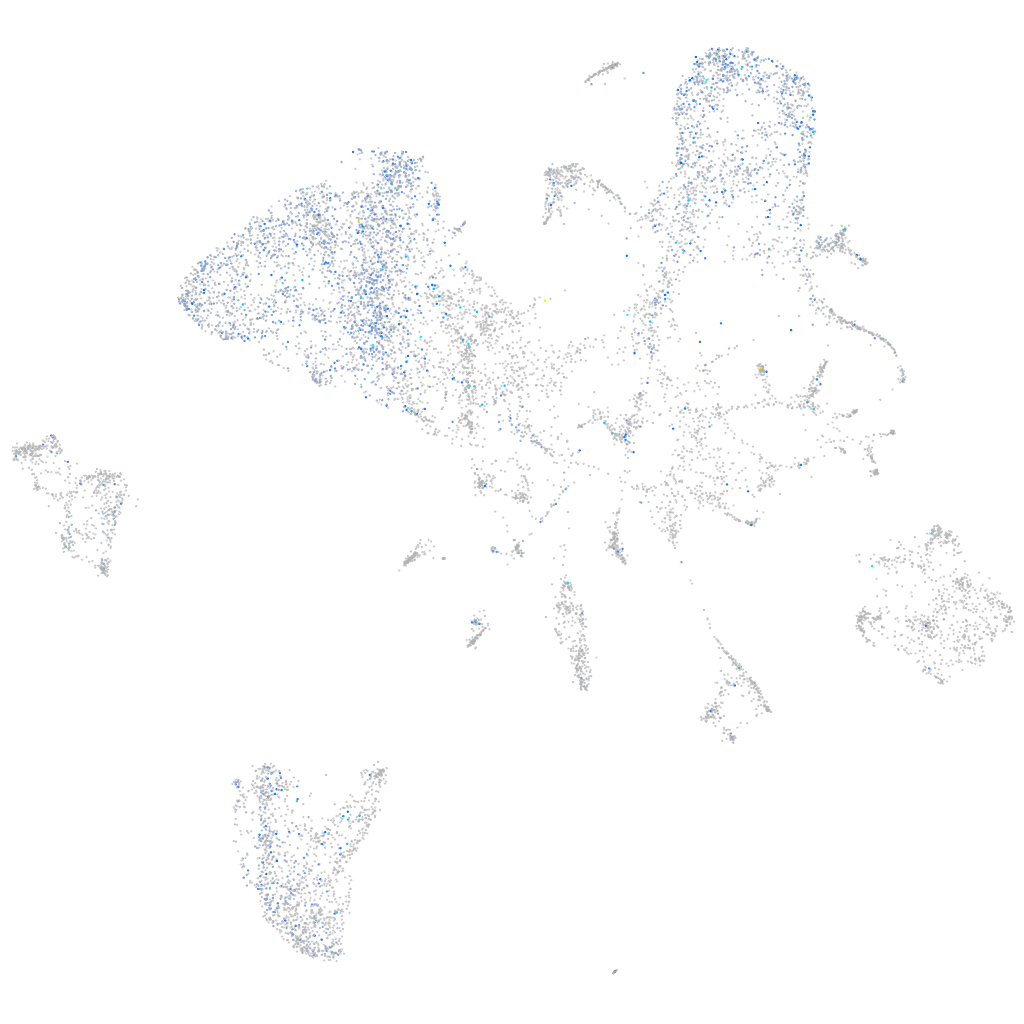
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glud1b | 0.267 | hmgn6 | -0.158 |
| rdh1 | 0.247 | si:ch211-222l21.1 | -0.154 |
| scp2a | 0.245 | si:ch73-1a9.3 | -0.148 |
| dhrs9 | 0.244 | marcksl1b | -0.147 |
| slc37a4a | 0.241 | hmgb3a | -0.146 |
| srd5a2a | 0.239 | h3f3d | -0.141 |
| slco1d1 | 0.237 | hmgb2a | -0.139 |
| apoa4b.1 | 0.236 | epcam | -0.138 |
| cx32.3 | 0.235 | jpt1b | -0.133 |
| gapdh | 0.235 | ywhaz | -0.131 |
| gpx4a | 0.234 | h2afvb | -0.130 |
| sod1 | 0.233 | ptmab | -0.129 |
| fbp1b | 0.233 | marcksb | -0.128 |
| g6pca.2 | 0.233 | hmgb1b | -0.127 |
| apobb.1 | 0.233 | tuba8l4 | -0.126 |
| cyp4v8 | 0.232 | hmgb1a | -0.126 |
| cyp8b1 | 0.230 | hmgn2 | -0.126 |
| dgat2 | 0.229 | ubc | -0.124 |
| eno3 | 0.228 | nucks1a | -0.124 |
| mgst1.2 | 0.228 | aldoaa | -0.124 |
| ugt1a7 | 0.228 | si:ch73-281n10.2 | -0.123 |
| sult2st2 | 0.227 | cx43.4 | -0.123 |
| apoc2 | 0.227 | khdrbs1a | -0.122 |
| ugt5b4 | 0.226 | syncrip | -0.120 |
| gcshb | 0.225 | hspb1 | -0.120 |
| sdr16c5b | 0.224 | cldnb | -0.119 |
| mdh1aa | 0.223 | eef1a1l1 | -0.119 |
| gstr | 0.222 | hdac1 | -0.118 |
| cat | 0.219 | hnrnpaba | -0.117 |
| suclg1 | 0.218 | tubb4b | -0.116 |
| abat | 0.218 | akap12b | -0.116 |
| abcc2 | 0.218 | cldn7b | -0.115 |
| aldh6a1 | 0.218 | hmgb2b | -0.115 |
| gstt1a | 0.217 | id1 | -0.114 |
| mttp | 0.217 | cd9b | -0.113 |