transmembrane protein 14Ca
ZFIN 
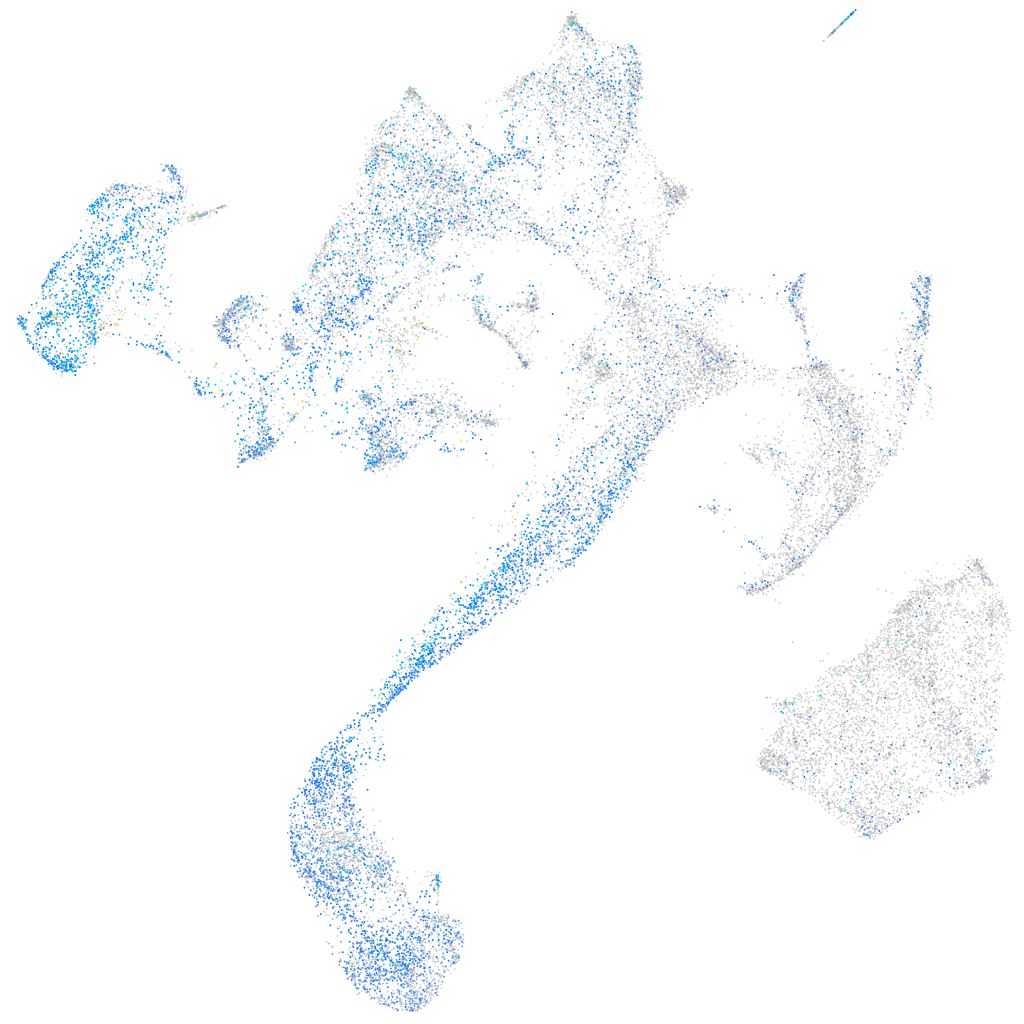
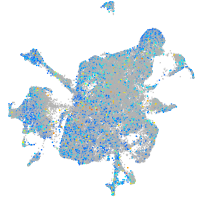
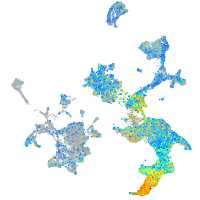
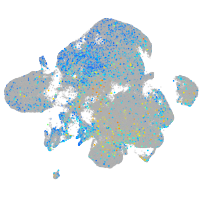
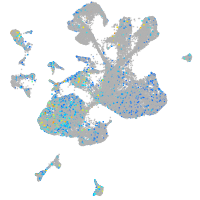
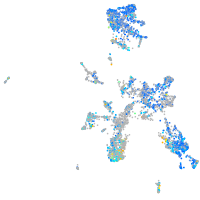
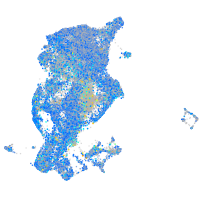
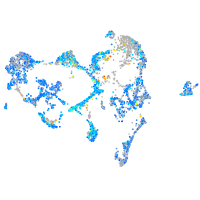
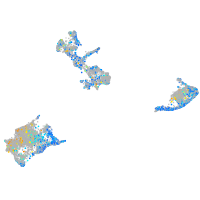
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5if1b | 0.396 | si:ch211-222l21.1 | -0.356 |
| aldoab | 0.391 | si:ch73-1a9.3 | -0.352 |
| atp2a1 | 0.377 | hsp90ab1 | -0.345 |
| tmem38a | 0.377 | si:ch73-281n10.2 | -0.345 |
| srl | 0.375 | ptmab | -0.341 |
| actc1b | 0.374 | hmgb2a | -0.339 |
| cox17 | 0.372 | pabpc1a | -0.336 |
| gapdh | 0.367 | hmgb2b | -0.334 |
| ckmb | 0.366 | h3f3d | -0.318 |
| mdh2 | 0.365 | hmga1a | -0.317 |
| ckma | 0.361 | cx43.4 | -0.314 |
| desma | 0.359 | hnrnpaba | -0.312 |
| ak1 | 0.359 | khdrbs1a | -0.308 |
| cav3 | 0.358 | cirbpa | -0.307 |
| pabpc4 | 0.355 | nucks1a | -0.306 |
| eno3 | 0.355 | cirbpb | -0.305 |
| suclg1 | 0.353 | h2afvb | -0.305 |
| casq2 | 0.351 | anp32b | -0.305 |
| atp5mc1 | 0.350 | seta | -0.301 |
| atp5meb | 0.348 | anp32a | -0.299 |
| ttn.2 | 0.348 | hmgn7 | -0.298 |
| chchd10 | 0.344 | syncrip | -0.297 |
| pgam2 | 0.344 | hnrnpabb | -0.292 |
| atp1a2a | 0.343 | si:ch211-288g17.3 | -0.289 |
| cox7a2a | 0.341 | tuba8l4 | -0.285 |
| ank1a | 0.340 | cbx3a | -0.284 |
| sdhdb | 0.338 | fthl27 | -0.283 |
| CABZ01072309.1 | 0.337 | ran | -0.280 |
| got2b | 0.337 | hnrnpa0b | -0.279 |
| atp5f1b | 0.337 | hmgn2 | -0.278 |
| tpi1b | 0.335 | ubc | -0.276 |
| CABZ01078594.1 | 0.333 | hmgn6 | -0.275 |
| pgk1 | 0.331 | marcksb | -0.269 |
| neb | 0.330 | sumo3a | -0.267 |
| nme2b.2 | 0.330 | ncl | -0.266 |