transmembrane protein 145
ZFIN 
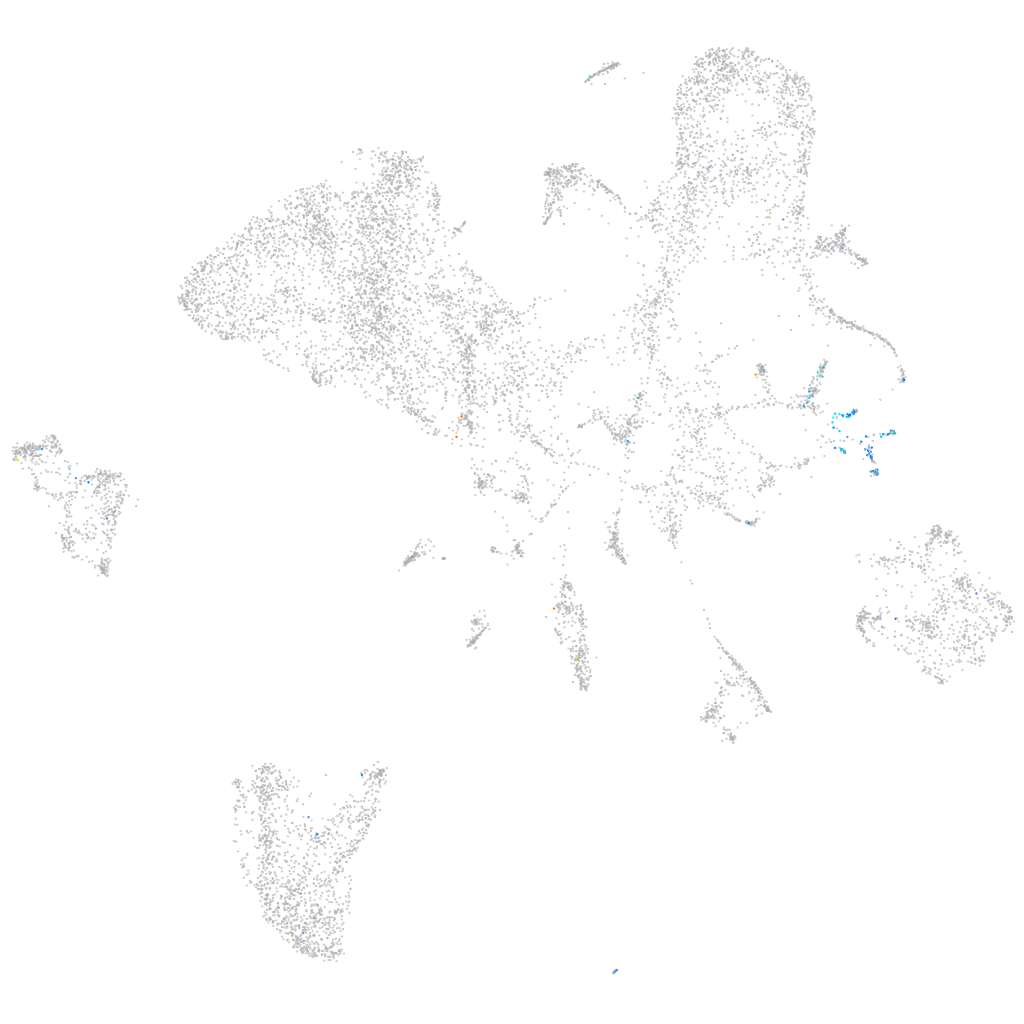
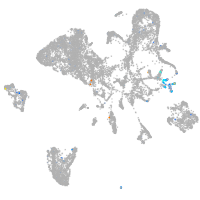
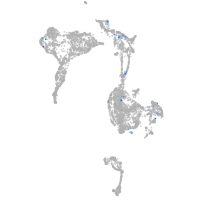
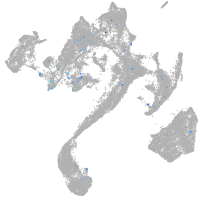
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.322 | gapdh | -0.093 |
| si:dkey-153k10.9 | 0.318 | ahcy | -0.092 |
| dkk3b | 0.318 | aldob | -0.084 |
| rprmb | 0.308 | rps20 | -0.083 |
| isl1 | 0.302 | gamt | -0.080 |
| camk1da | 0.283 | gstt1a | -0.077 |
| SLC5A10 | 0.283 | eno3 | -0.076 |
| kcnj11 | 0.278 | fbp1b | -0.074 |
| tspan7b | 0.275 | aldh6a1 | -0.074 |
| gcgb | 0.273 | fabp3 | -0.072 |
| scg2a | 0.273 | cox6b1 | -0.070 |
| si:ch73-287m6.1 | 0.268 | mat1a | -0.070 |
| pcsk1 | 0.263 | gstr | -0.070 |
| pcsk1nl | 0.262 | cx32.3 | -0.069 |
| neurod1 | 0.259 | apoa4b.1 | -0.069 |
| pcsk2 | 0.258 | hspe1 | -0.068 |
| pax6b | 0.256 | ppa1b | -0.068 |
| slc35g2b | 0.253 | glud1b | -0.067 |
| slc1a4 | 0.253 | apoc2 | -0.067 |
| LOC571123 | 0.249 | si:dkey-16p21.8 | -0.067 |
| ndufa4l2a | 0.247 | agxta | -0.066 |
| CR774179.5 | 0.246 | apoa1b | -0.066 |
| syt1a | 0.245 | gnmt | -0.066 |
| cadm2a | 0.243 | sult2st2 | -0.066 |
| scg3 | 0.243 | nupr1b | -0.065 |
| insm1a | 0.239 | scp2a | -0.065 |
| disp2 | 0.239 | bhmt | -0.064 |
| pcdh20 | 0.237 | aqp12 | -0.064 |
| sst2 | 0.235 | gsta.1 | -0.064 |
| cacna2d2a | 0.231 | rmdn1 | -0.063 |
| abhd15a | 0.231 | gcshb | -0.063 |
| cpe | 0.231 | grhprb | -0.062 |
| kcnk9 | 0.230 | agxtb | -0.062 |
| vamp2 | 0.227 | aldh7a1 | -0.062 |
| cacna1c | 0.227 | dhdhl | -0.062 |