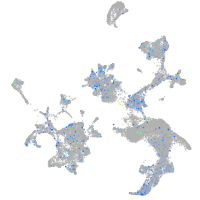
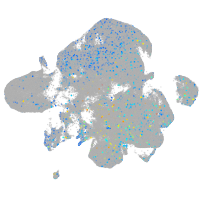
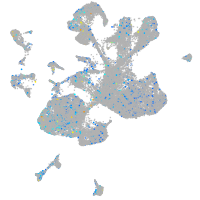
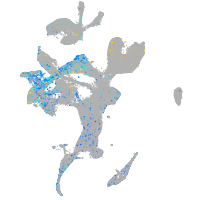
transmembrane protein 135
ZFIN 
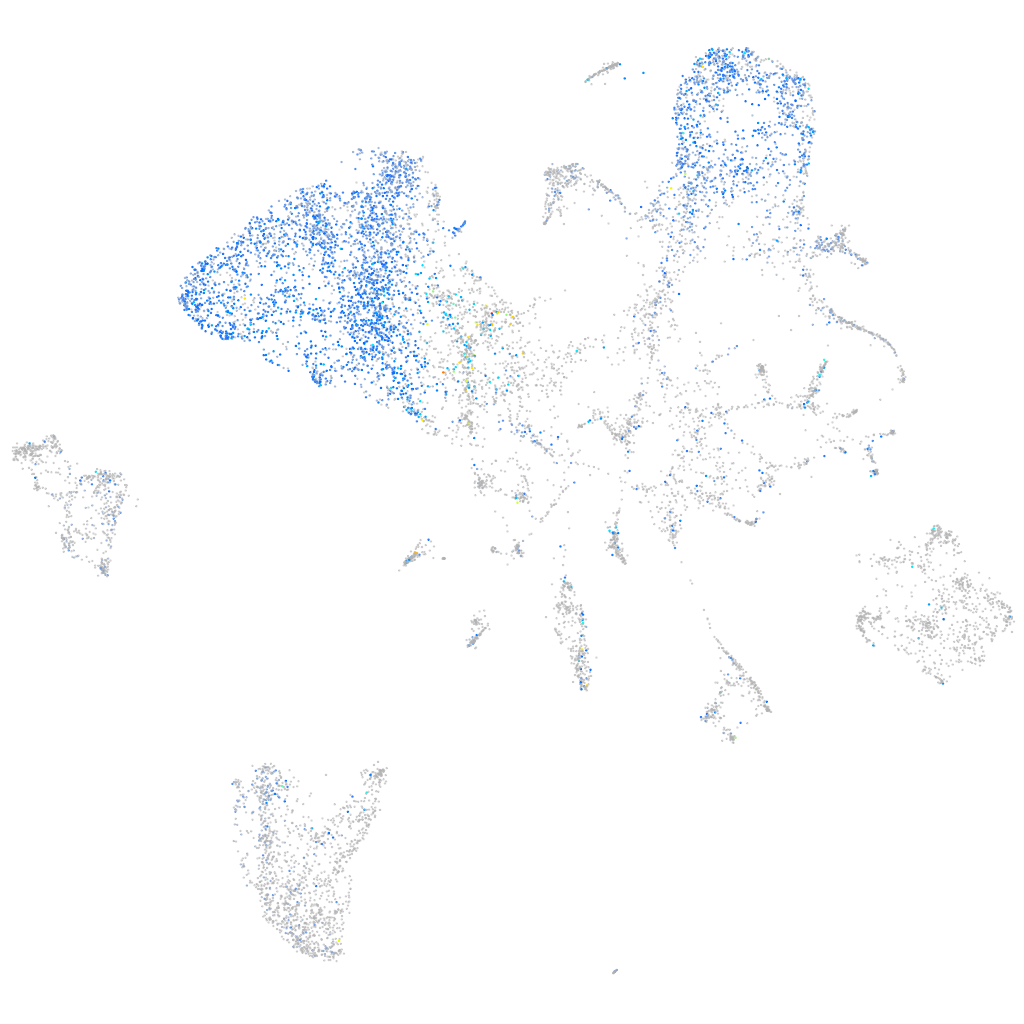
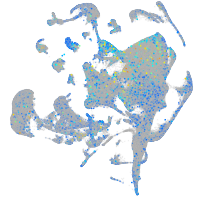
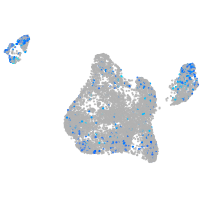
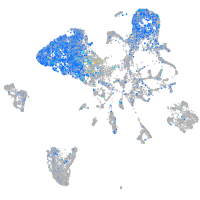
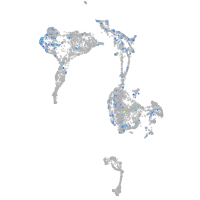
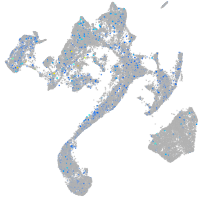
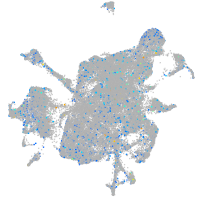
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scp2a | 0.515 | si:ch211-222l21.1 | -0.275 |
| apoa4b.1 | 0.495 | epcam | -0.261 |
| haao | 0.479 | hmgn6 | -0.253 |
| mgst1.2 | 0.474 | slc38a5b | -0.250 |
| msrb2 | 0.472 | hmgb3a | -0.242 |
| gpx4a | 0.470 | h3f3d | -0.242 |
| suclg1 | 0.469 | idh2 | -0.241 |
| glud1b | 0.468 | si:ch73-1a9.3 | -0.234 |
| mdh1aa | 0.468 | ubc | -0.227 |
| tdo2a | 0.464 | rtn1a | -0.220 |
| pgam1a | 0.464 | ywhaz | -0.219 |
| apoc2 | 0.463 | hmgb2a | -0.217 |
| rdh1 | 0.462 | marcksl1b | -0.217 |
| igfbp2a | 0.460 | hmgn2 | -0.217 |
| apoa1b | 0.458 | tuba8l4 | -0.216 |
| cyp4v8 | 0.458 | hmgb1a | -0.216 |
| slc37a4a | 0.456 | jpt1b | -0.216 |
| fbp1b | 0.455 | marcksb | -0.215 |
| gapdh | 0.455 | hmgb1b | -0.214 |
| acaa2 | 0.455 | eef1a1l1 | -0.212 |
| etnppl | 0.454 | glulb | -0.211 |
| pgm1 | 0.453 | syncrip | -0.211 |
| aldh1l1 | 0.453 | cldnb | -0.211 |
| srd5a2a | 0.452 | khdrbs1a | -0.209 |
| cyp8b1 | 0.452 | aldoaa | -0.206 |
| slco1d1 | 0.451 | cd9b | -0.203 |
| gstt1a | 0.446 | pfn1 | -0.201 |
| afp4 | 0.446 | her9 | -0.198 |
| abat | 0.444 | hnrnpaba | -0.198 |
| pgk1 | 0.444 | h2afvb | -0.197 |
| g6pca.2 | 0.443 | si:ch211-195b11.3 | -0.197 |
| upb1 | 0.443 | cldn7b | -0.197 |
| pygl | 0.443 | cirbpa | -0.197 |
| ugt5b4 | 0.441 | acin1a | -0.197 |
| shbg | 0.440 | si:dkey-42i9.4 | -0.196 |