tight junction protein 1b
ZFIN 
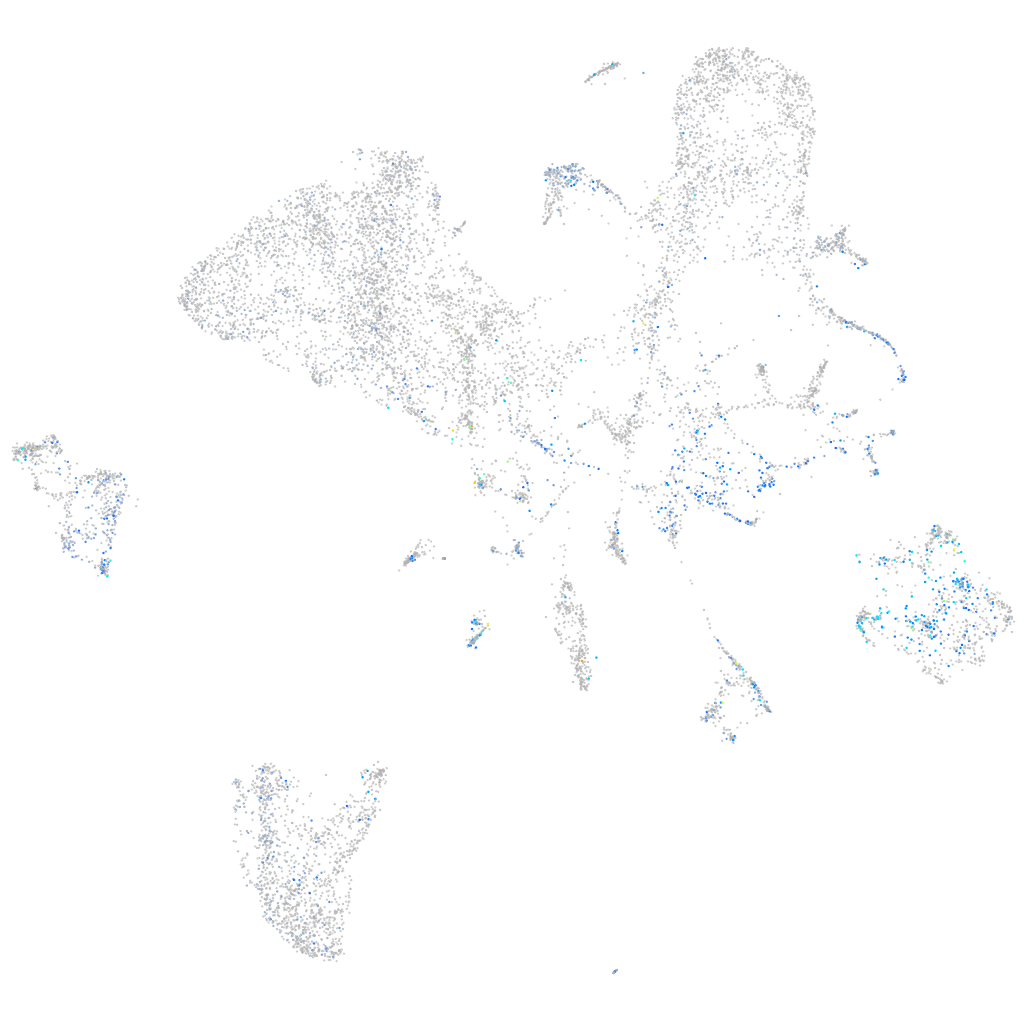
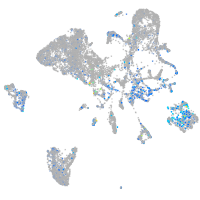
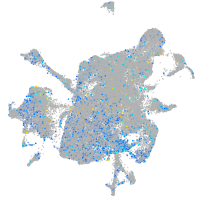
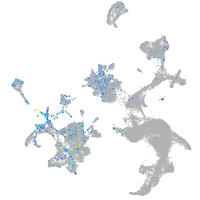
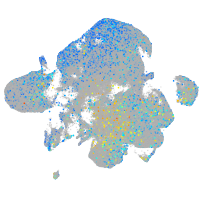
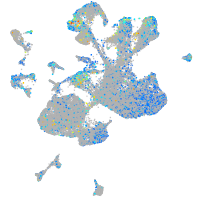
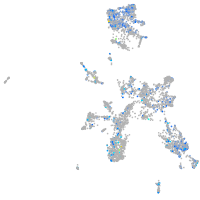
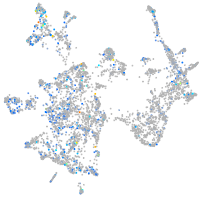
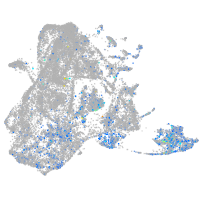
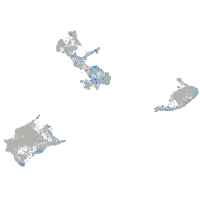
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| qkia | 0.236 | gapdh | -0.210 |
| cdh6 | 0.230 | ahcy | -0.208 |
| akap12b | 0.227 | gamt | -0.198 |
| marcksb | 0.219 | sod1 | -0.179 |
| nucks1a | 0.211 | eno3 | -0.178 |
| hspb1 | 0.209 | apoc2 | -0.178 |
| hnrnpa1a | 0.209 | gpx4a | -0.177 |
| pltp | 0.208 | fbp1b | -0.175 |
| marcksl1b | 0.207 | apoa4b.1 | -0.173 |
| hp1bp3 | 0.206 | gatm | -0.170 |
| acin1a | 0.204 | suclg1 | -0.169 |
| hmgn6 | 0.204 | mdh1aa | -0.168 |
| si:ch211-152c2.3 | 0.199 | scp2a | -0.166 |
| lima1a | 0.196 | gstt1a | -0.164 |
| syncrip | 0.195 | afp4 | -0.164 |
| nr6a1a | 0.195 | prdx2 | -0.163 |
| anp32a | 0.194 | aldob | -0.162 |
| hmgb3a | 0.193 | apoa1b | -0.160 |
| asph | 0.192 | tpi1b | -0.160 |
| si:ch73-281n10.2 | 0.192 | pklr | -0.157 |
| si:ch73-1a9.3 | 0.192 | dap | -0.156 |
| cx43.4 | 0.191 | glud1b | -0.156 |
| apela | 0.191 | atp5mc1 | -0.151 |
| tgif1 | 0.190 | rpl37 | -0.150 |
| ilf3b | 0.189 | rdh1 | -0.149 |
| smc1al | 0.186 | sult2st2 | -0.149 |
| ctnnb1 | 0.185 | mat1a | -0.148 |
| id1 | 0.185 | srd5a2a | -0.146 |
| smo | 0.184 | suclg2 | -0.143 |
| hnrnpa1b | 0.183 | pnp4b | -0.143 |
| tpm4a | 0.183 | pla2g12b | -0.143 |
| cirbpa | 0.183 | bhmt | -0.143 |
| NC-002333.4 | 0.183 | apobb.1 | -0.143 |
| smarca4a | 0.183 | cox7a1 | -0.142 |
| khdrbs1a | 0.183 | nipsnap3a | -0.142 |