tight junction protein 1a
ZFIN 
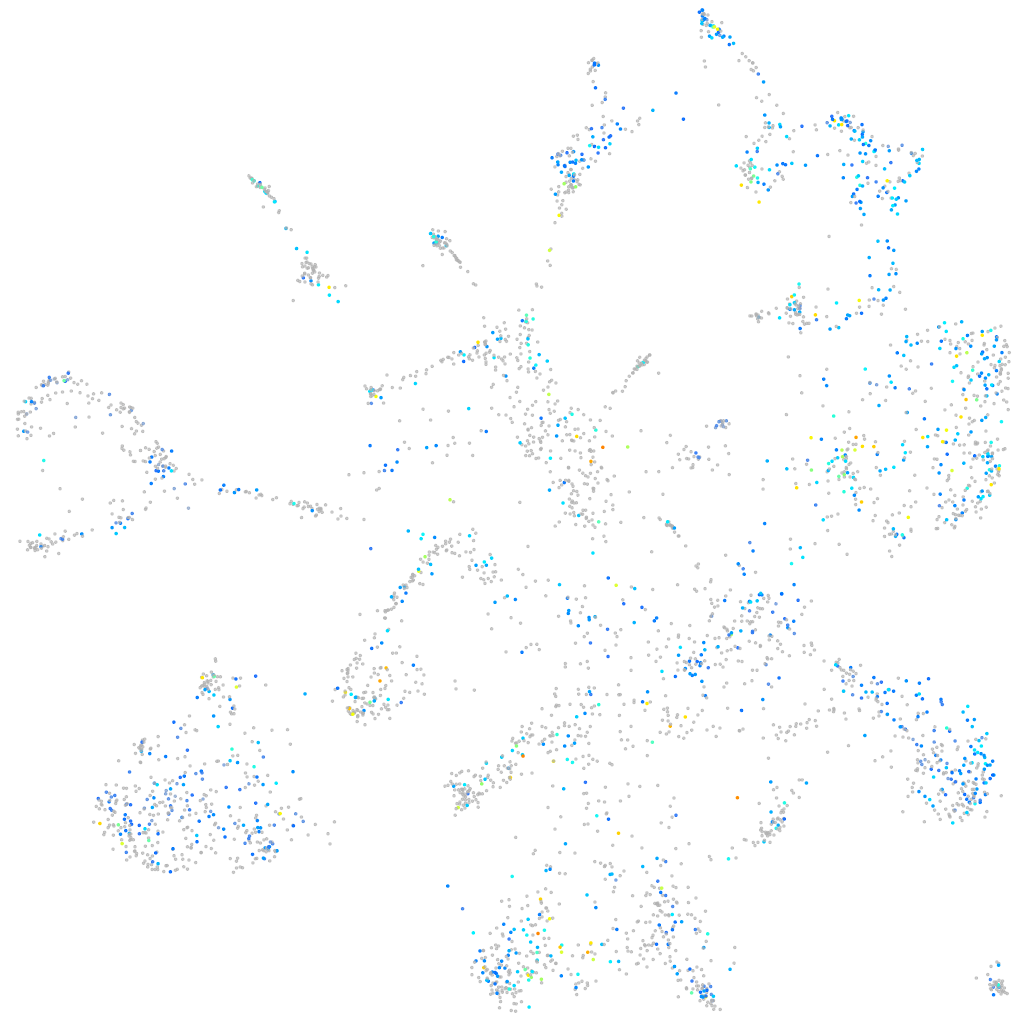
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.207 | gapdhs | -0.098 |
| rhag | 0.188 | ak1 | -0.087 |
| tmem88b | 0.185 | BX088707.3 | -0.087 |
| jam2b | 0.172 | tpi1b | -0.086 |
| krt8 | 0.169 | eno1a | -0.080 |
| krt94 | 0.167 | ldha | -0.078 |
| tmem98 | 0.162 | aldocb | -0.074 |
| cd151 | 0.160 | ptmaa | -0.074 |
| cavin1b | 0.158 | tpm1 | -0.073 |
| bnc2 | 0.156 | igfbp7 | -0.072 |
| cavin2b | 0.155 | tdh | -0.072 |
| ftr82 | 0.153 | pgk1 | -0.072 |
| tgm2b | 0.153 | ndufa4l2a | -0.071 |
| plp2 | 0.153 | cx43 | -0.071 |
| COLEC10 | 0.152 | pkma | -0.071 |
| cxadr | 0.152 | acta2 | -0.068 |
| cav1 | 0.150 | inka1a | -0.068 |
| aqp1a.1 | 0.148 | agtr2 | -0.067 |
| tjp2b | 0.146 | fbp2 | -0.066 |
| akap12b | 0.145 | si:ch211-270g19.5 | -0.065 |
| alx4a | 0.144 | pgam1a | -0.064 |
| bcam | 0.144 | eno3 | -0.064 |
| thy1 | 0.142 | mamdc2a | -0.063 |
| krt18a.1 | 0.142 | lbh | -0.063 |
| metrnlb | 0.139 | foxl1 | -0.063 |
| gstm.3 | 0.139 | pvalb2 | -0.061 |
| phactr4a | 0.139 | barx1 | -0.060 |
| cdon | 0.138 | inhbaa | -0.060 |
| mdka | 0.135 | foxf2b | -0.060 |
| eppk1 | 0.135 | pfkpa | -0.060 |
| gata5 | 0.135 | bgnb | -0.059 |
| scarb2a | 0.135 | chac1 | -0.059 |
| fabp11a | 0.134 | timm8a | -0.058 |
| colec11 | 0.132 | vwde | -0.058 |
| mmel1 | 0.131 | hoxa10b | -0.058 |