TIMP metallopeptidase inhibitor 2b
ZFIN 
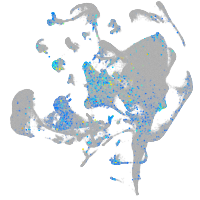
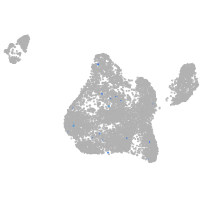
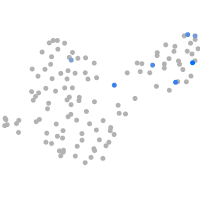
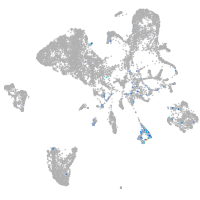
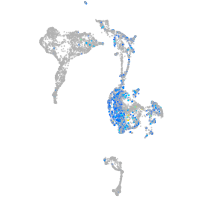
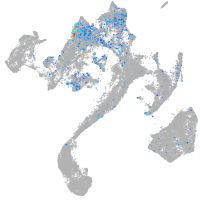
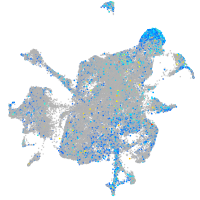
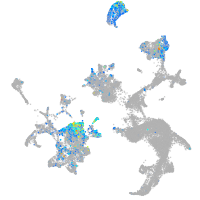
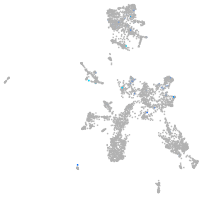
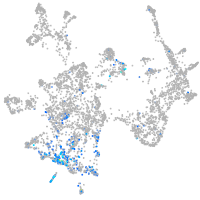
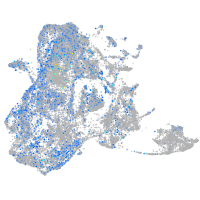
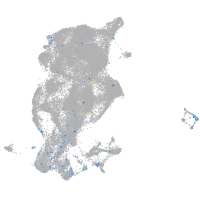
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01055365.1 | 0.362 | gapdh | -0.115 |
| tgfb3 | 0.352 | fbp1b | -0.114 |
| pmp22b | 0.350 | gamt | -0.110 |
| sim1b | 0.345 | aldob | -0.110 |
| mxra8b | 0.322 | gpx4a | -0.109 |
| f3a | 0.321 | gatm | -0.108 |
| cabp2b | 0.321 | ahcy | -0.104 |
| marcksl1a | 0.320 | apoa4b.1 | -0.099 |
| anxa3b | 0.315 | apoc2 | -0.099 |
| sftpba | 0.303 | scp2a | -0.097 |
| ctgfa | 0.302 | mdh1aa | -0.097 |
| cav1 | 0.301 | mat1a | -0.097 |
| spock3 | 0.300 | pklr | -0.095 |
| gna15.1 | 0.297 | apoa1b | -0.095 |
| arhgap29b | 0.297 | sod1 | -0.094 |
| cd151 | 0.292 | afp4 | -0.094 |
| abca12 | 0.291 | gstt1a | -0.092 |
| lama5 | 0.290 | bhmt | -0.092 |
| rab32a | 0.288 | sult2st2 | -0.090 |
| spns2 | 0.285 | rdh1 | -0.087 |
| bcam | 0.276 | si:dkey-16p21.8 | -0.087 |
| tekt3 | 0.275 | pnp4b | -0.086 |
| b3gnt5a | 0.274 | agxtb | -0.086 |
| anxa13 | 0.274 | dap | -0.086 |
| sgms1 | 0.273 | abat | -0.085 |
| erbb3a | 0.272 | gcshb | -0.084 |
| krt94 | 0.272 | slco1d1 | -0.084 |
| ceacam1 | 0.270 | apobb.1 | -0.084 |
| hspa12b | 0.267 | tdo2a | -0.084 |
| cav2 | 0.262 | haao | -0.083 |
| vwa1 | 0.262 | tpi1b | -0.083 |
| cygb1 | 0.260 | aldh6a1 | -0.083 |
| capn2a | 0.258 | suclg2 | -0.083 |
| slc6a14 | 0.254 | gpd1b | -0.083 |
| sparc | 0.252 | nipsnap3a | -0.081 |