thymocyte nuclear protein 1
ZFIN 
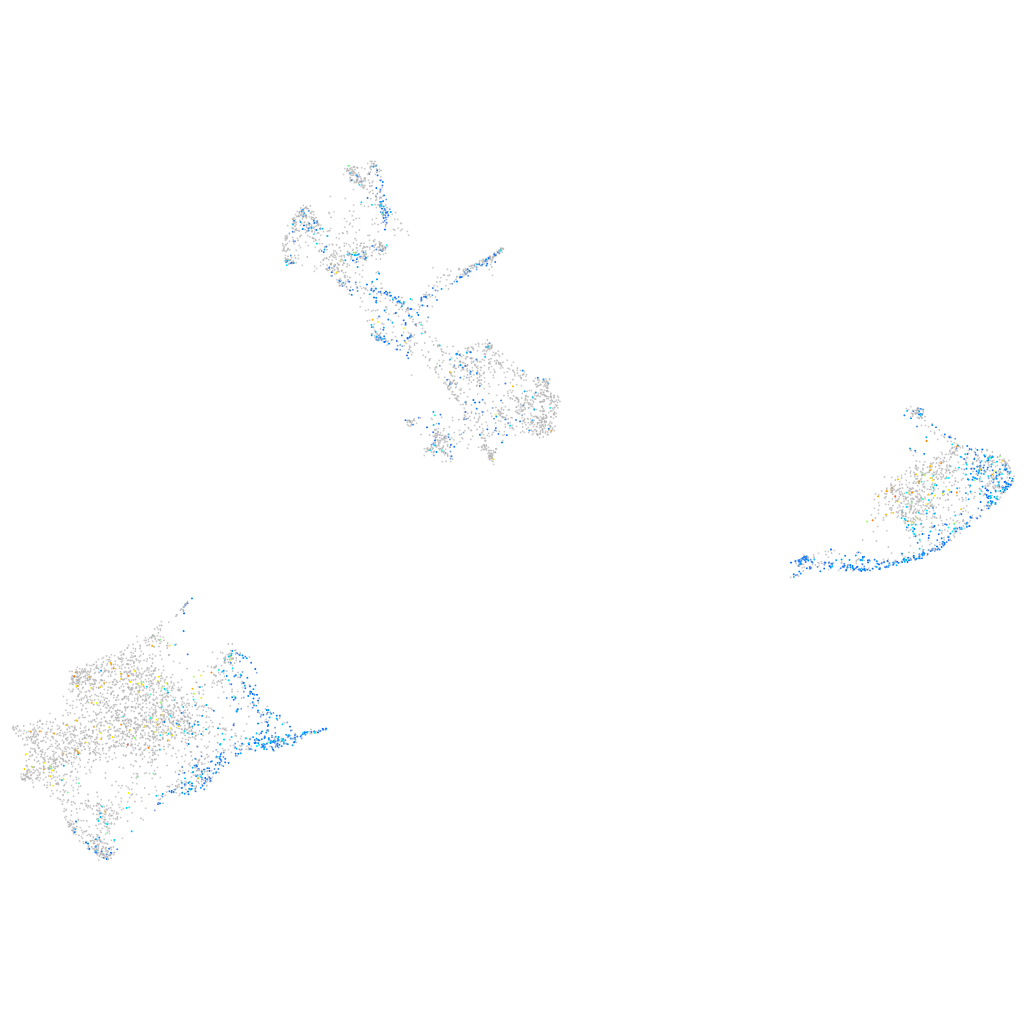
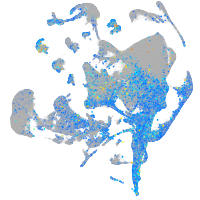
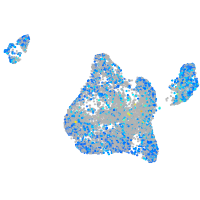
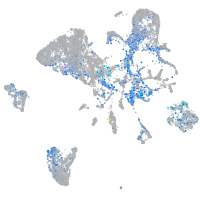
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ptges3b | 0.196 | pvalb1 | -0.106 |
| rab32a | 0.184 | pvalb2 | -0.100 |
| npm1a | 0.184 | nova2 | -0.098 |
| tmem243b | 0.183 | tmsb | -0.098 |
| sypl2b | 0.178 | actc1b | -0.098 |
| mitfa | 0.177 | ptmaa | -0.092 |
| gpr143 | 0.177 | gpm6aa | -0.090 |
| naga | 0.172 | tuba1c | -0.090 |
| rabl6b | 0.170 | hbae3 | -0.089 |
| hspd1 | 0.168 | elavl3 | -0.085 |
| tspan36 | 0.168 | uraha | -0.085 |
| slc37a2 | 0.167 | CABZ01021592.1 | -0.084 |
| atp6v1g1 | 0.167 | hbbe1.1 | -0.084 |
| uqcrh | 0.161 | sncb | -0.082 |
| qdpra | 0.160 | si:dkey-251i10.2 | -0.080 |
| snx2 | 0.159 | ndrg2 | -0.079 |
| abracl | 0.159 | hbbe1.3 | -0.077 |
| tfap2e | 0.157 | zc4h2 | -0.077 |
| syngr1a | 0.157 | si:ch211-251b21.1 | -0.075 |
| scpep1 | 0.156 | mylz3 | -0.074 |
| degs1 | 0.155 | stmn1b | -0.073 |
| psap | 0.155 | gng3 | -0.072 |
| xbp1 | 0.154 | myhz1.1 | -0.071 |
| ap3m2 | 0.154 | sox11a | -0.070 |
| slc30a7 | 0.153 | tnni2a.4 | -0.069 |
| cst14b.1 | 0.153 | epb41a | -0.068 |
| bace2 | 0.152 | mdh1aa | -0.068 |
| SPAG9 | 0.152 | krtt1c19e | -0.067 |
| slc45a2 | 0.152 | mylpfa | -0.067 |
| slc3a2a | 0.152 | CU467822.1 | -0.066 |
| ssr2 | 0.151 | apoa2 | -0.066 |
| rab38 | 0.151 | sesn1 | -0.065 |
| rnaseka | 0.151 | tnnt3b | -0.065 |
| tyr | 0.151 | ebf3a | -0.065 |
| si:zfos-943e10.1 | 0.150 | id4 | -0.065 |