transferrin receptor 2
ZFIN 
Other cell groups


all cells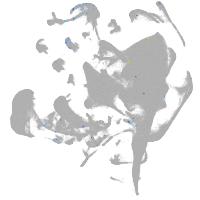 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 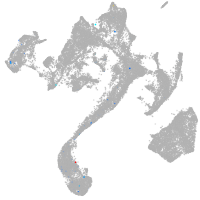 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 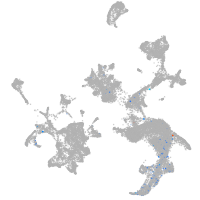 |
pronephros 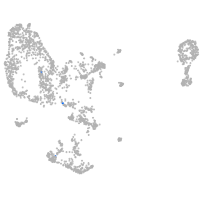 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cpb2 | 0.138 | hmgb1b | -0.029 |
| c8a | 0.134 | hmgb3a | -0.029 |
| si:ch211-170d8.5 | 0.134 | ckbb | -0.028 |
| a2ml | 0.132 | marcksl1a | -0.028 |
| serpinf2a | 0.132 | marcksb | -0.027 |
| si:ch211-186e20.7 | 0.132 | nucks1a | -0.027 |
| c3a.3 | 0.131 | tuba1c | -0.027 |
| si:ch211-288g17.4 | 0.131 | gpm6aa | -0.026 |
| cfb | 0.130 | jpt1b | -0.026 |
| hbxb | 0.130 | nova2 | -0.026 |
| abcb11b | 0.129 | gnb1b | -0.025 |
| c3a.6 | 0.129 | hnrnpa0a | -0.025 |
| g6pca.1 | 0.129 | si:ch211-288g17.3 | -0.025 |
| masp2 | 0.129 | acin1a | -0.023 |
| zgc:112265 | 0.129 | atrx | -0.023 |
| c3a.1 | 0.128 | gapdhs | -0.023 |
| hal | 0.128 | gpm6ab | -0.023 |
| shbg | 0.128 | hp1bp3 | -0.023 |
| vtna | 0.128 | mdkb | -0.023 |
| f7 | 0.128 | rnasekb | -0.023 |
| agt | 0.127 | si:dkey-276j7.1 | -0.023 |
| c3a.2 | 0.127 | ywhaqb | -0.023 |
| cfhl4 | 0.127 | zc4h2 | -0.023 |
| f7i | 0.127 | atp6v0cb | -0.022 |
| f9b | 0.127 | chd4a | -0.022 |
| c9 | 0.126 | h2afy2 | -0.022 |
| cfhl1 | 0.126 | si:ch211-137a8.4 | -0.022 |
| f5 | 0.126 | si:ch73-46j18.5 | -0.022 |
| fga | 0.126 | stmn1b | -0.022 |
| itih2 | 0.126 | tuba1a | -0.022 |
| oxct1b | 0.126 | tubb5 | -0.022 |
| proca | 0.126 | CABZ01080702.1 | -0.021 |
| serpinc1 | 0.126 | elavl3 | -0.021 |
| serpind1 | 0.126 | hdac1 | -0.021 |
| apoba | 0.125 | hnrnpa1a | -0.021 |




