"testis expressed 264, ER-phagy receptor a"
ZFIN 
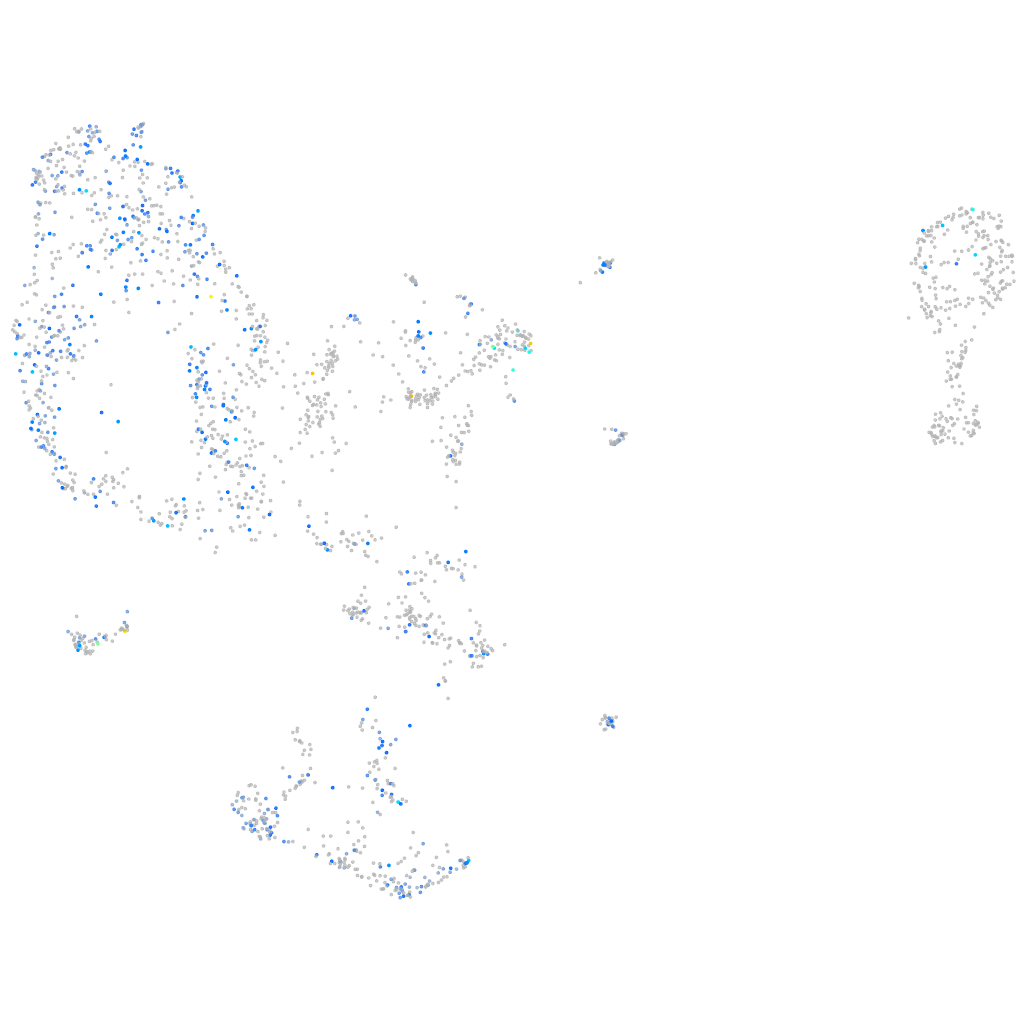
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lrrtm4l2 | 0.198 | hmgb2a | -0.175 |
| ndfip1 | 0.198 | marcksb | -0.168 |
| mt-atp6 | 0.197 | si:ch73-1a9.3 | -0.162 |
| mt-nd1 | 0.197 | hmgb2b | -0.161 |
| aldh7a1 | 0.191 | hspb1 | -0.161 |
| pttg1ipb | 0.189 | wu:fb97g03 | -0.154 |
| eps8l3b | 0.187 | zgc:110425 | -0.153 |
| gpx4a | 0.185 | si:ch73-281n10.2 | -0.152 |
| slc26a1 | 0.183 | hmga1a | -0.150 |
| CABZ01044023.1 | 0.182 | fbl | -0.149 |
| bsg | 0.182 | cirbpa | -0.148 |
| pllp | 0.181 | apoeb | -0.138 |
| eif4a1b | 0.181 | cbx3a | -0.134 |
| si:ch1073-443f11.2 | 0.180 | hmgn2 | -0.131 |
| elovl1b | 0.180 | nucks1a | -0.130 |
| gcshb | 0.180 | si:ch211-288g17.3 | -0.128 |
| cygb2 | 0.179 | anp32b | -0.128 |
| igfbp5b | 0.178 | sumo3b | -0.128 |
| npc2 | 0.178 | syncrip | -0.128 |
| atp8a2 | 0.177 | lig1 | -0.128 |
| scpep1 | 0.177 | hnrnpabb | -0.128 |
| cdh17 | 0.177 | nop58 | -0.127 |
| alpl | 0.177 | znfl2a | -0.126 |
| gapdh | 0.176 | seta | -0.124 |
| tmem266 | 0.175 | nasp | -0.123 |
| aldh6a1 | 0.175 | ebna1bp2 | -0.123 |
| fuca1.1 | 0.173 | ppig | -0.123 |
| mt-co2 | 0.173 | si:ch211-222l21.1 | -0.123 |
| trim101 | 0.173 | msna | -0.122 |
| ctsd | 0.173 | snrpd3l | -0.121 |
| COX3 | 0.172 | h2afvb | -0.121 |
| prdx2 | 0.171 | apoc1 | -0.120 |
| akt2l | 0.171 | lmo2 | -0.120 |
| acsf2 | 0.171 | brd3a | -0.119 |
| gstk1 | 0.171 | stmn1a | -0.119 |