tet methylcytosine dioxygenase 3
ZFIN 
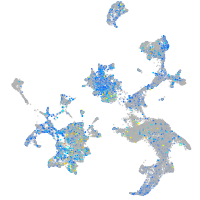
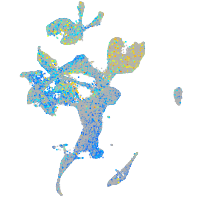
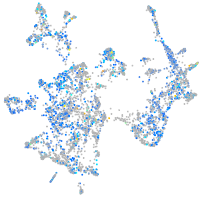
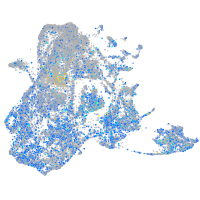
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.224 | gstp1 | -0.124 |
| nova2 | 0.218 | si:dkey-251i10.2 | -0.124 |
| zc4h2 | 0.207 | slc22a7a | -0.120 |
| chd4a | 0.205 | gch2 | -0.118 |
| gpm6aa | 0.201 | zgc:110339 | -0.114 |
| tmeff1b | 0.193 | uraha | -0.111 |
| si:ch211-222l21.1 | 0.192 | actb2 | -0.109 |
| tuba1c | 0.190 | CABZ01021592.1 | -0.106 |
| ptmab | 0.189 | mdh1aa | -0.100 |
| stmn1b | 0.189 | tmem130 | -0.098 |
| hmgb1b | 0.185 | paics | -0.093 |
| marcksb | 0.182 | TMEM19 | -0.091 |
| ptmaa | 0.182 | akr1b1 | -0.091 |
| hmgb3a | 0.180 | phyhd1 | -0.091 |
| tmpob | 0.179 | pfn1 | -0.090 |
| myt1a | 0.179 | pts | -0.085 |
| rtn1a | 0.179 | cyb5a | -0.085 |
| cd99l2 | 0.178 | krt18b | -0.083 |
| rnasekb | 0.178 | aldob | -0.080 |
| marcksl1a | 0.177 | gpd1b | -0.079 |
| epb41a | 0.177 | pvalb2 | -0.077 |
| hnrnpa0b | 0.175 | pvalb1 | -0.072 |
| tuba1a | 0.174 | dio3a | -0.068 |
| celf4 | 0.174 | prdx5 | -0.067 |
| gng3 | 0.173 | reep5 | -0.067 |
| stx1b | 0.173 | krt8 | -0.066 |
| gng2 | 0.172 | aldh1l1 | -0.065 |
| ncam1a | 0.172 | aqp3a | -0.065 |
| si:ch211-288g17.3 | 0.171 | actc1b | -0.063 |
| fam168a | 0.170 | bco1 | -0.063 |
| si:ch73-1a9.3 | 0.169 | prdx1 | -0.060 |
| ankrd12 | 0.169 | cmtm3 | -0.060 |
| hnrnpaba | 0.169 | impdh1b | -0.058 |
| tmem35 | 0.168 | glrx | -0.058 |
| tmsb | 0.167 | mylpfa | -0.056 |