"thymocyte expressed, positive selection associated 1"
ZFIN 
Other cell groups


all cells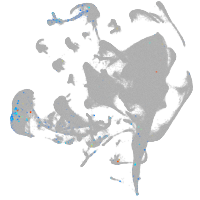 |
blastomeres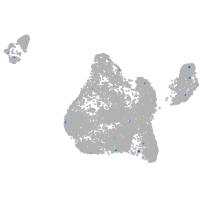 |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
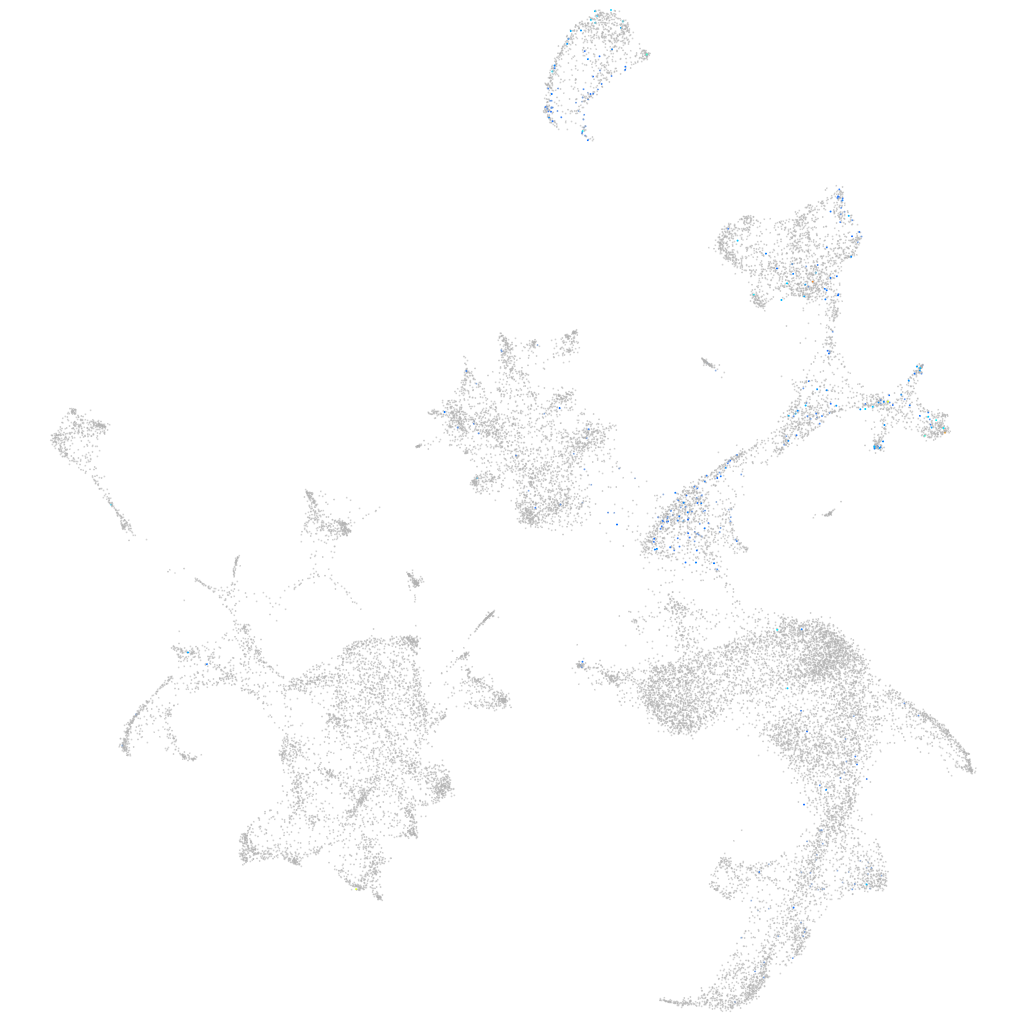
hematopoietic / vasculature 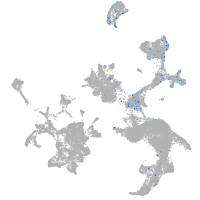 |
pronephros 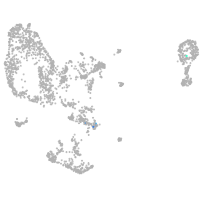 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm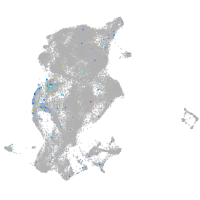 |
fin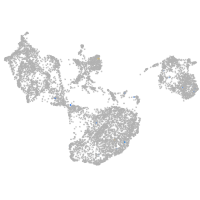 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dub | 0.129 | hbae3 | -0.065 |
| coro1a | 0.125 | hbbe1.3 | -0.062 |
| wasa | 0.123 | hbae1.3 | -0.056 |
| grap2b | 0.123 | si:ch211-250g4.3 | -0.055 |
| laptm5 | 0.121 | hbbe1.1 | -0.055 |
| itgae.2 | 0.120 | krt18a.1 | -0.054 |
| pfn1 | 0.115 | hbbe2 | -0.052 |
| rac2 | 0.113 | creg1 | -0.052 |
| ccr9a | 0.112 | hbae1.1 | -0.051 |
| syk | 0.112 | hbbe1.2 | -0.050 |
| LOC103909115 | 0.112 | aqp1a.1 | -0.050 |
| hmha1b | 0.111 | cahz | -0.050 |
| si:ch211-134a4.2 | 0.111 | tpm4a | -0.046 |
| lpar5a | 0.109 | cdh5 | -0.045 |
| myb | 0.109 | si:ch211-156j16.1 | -0.045 |
| cebpb | 0.109 | krt8 | -0.044 |
| rgs13 | 0.108 | egfl7 | -0.044 |
| si:ch211-214p16.1 | 0.107 | cpn1 | -0.043 |
| si:ch73-208g10.1 | 0.106 | nt5c2l1 | -0.043 |
| glipr1a | 0.106 | alas2 | -0.043 |
| fam49ba | 0.105 | tuba8l3 | -0.043 |
| arpc1b | 0.105 | cldn5b | -0.043 |
| si:dkey-13m1.2 | 0.103 | clec14a | -0.042 |
| si:dkey-262k9.4 | 0.103 | id1 | -0.042 |
| tnfaip8l2a | 0.102 | akap12b | -0.042 |
| itgb2 | 0.100 | si:dkeyp-97a10.2 | -0.042 |
| parvg | 0.100 | pmp22a | -0.041 |
| zgc:64051 | 0.100 | ctsla | -0.041 |
| tagapb | 0.100 | ecscr | -0.041 |
| fermt3b | 0.100 | ramp2 | -0.041 |
| ccdc88b | 0.099 | sox7 | -0.040 |
| LOC101883802 | 0.099 | marcksl1b | -0.040 |
| CABZ01033205.1 | 0.098 | tmem88a | -0.040 |
| arhgap15 | 0.097 | plvapb | -0.040 |
| fmnl1a | 0.097 | fstl1b | -0.040 |