teneurin transmembrane protein 3
ZFIN 
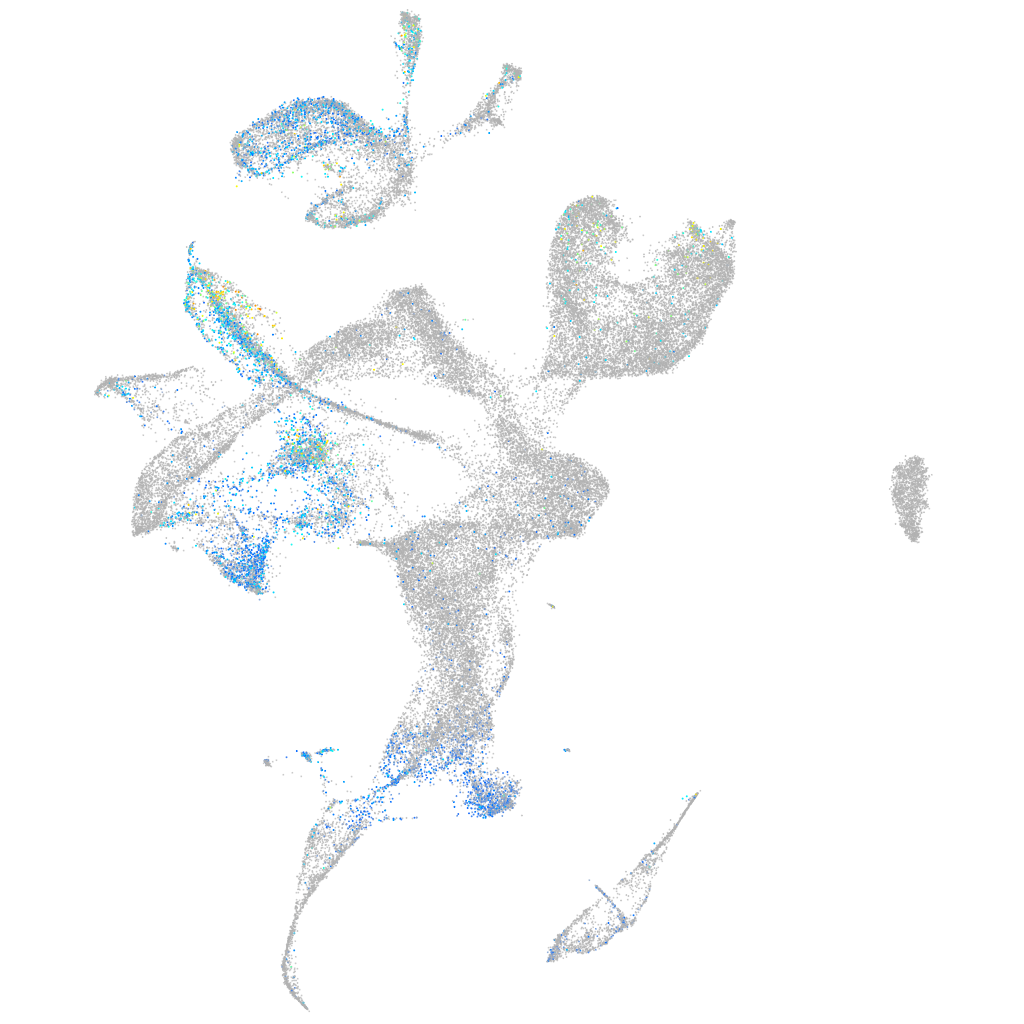
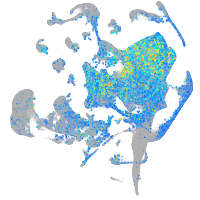
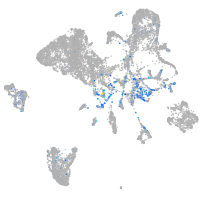
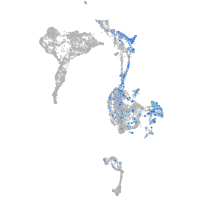
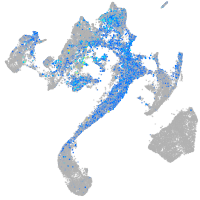
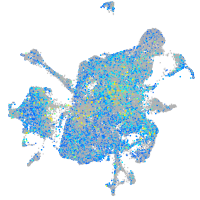
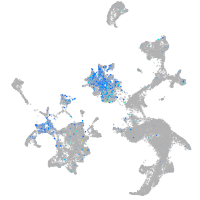
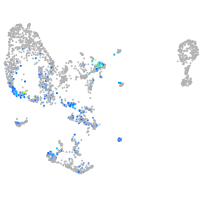
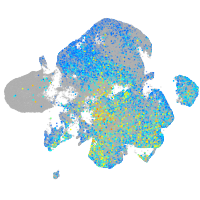
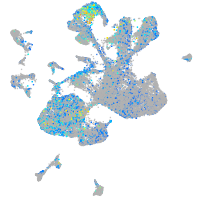
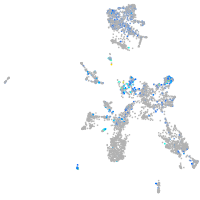
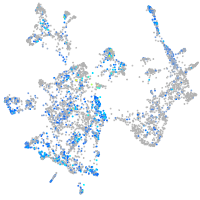
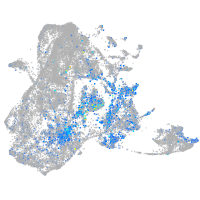
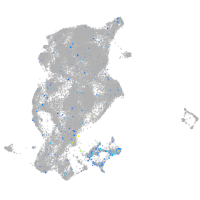
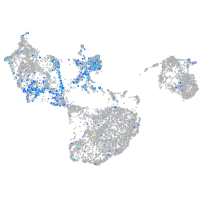
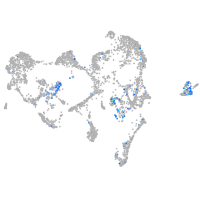
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.259 | hmgb2a | -0.181 |
| zfhx3 | 0.250 | fabp7a | -0.114 |
| rtn1b | 0.228 | neurod4 | -0.105 |
| islr2 | 0.224 | anp32e | -0.103 |
| stmn2a | 0.220 | hmga1a | -0.091 |
| tmsb | 0.209 | crx | -0.091 |
| elavl4 | 0.205 | rrm1 | -0.090 |
| rbpms2a | 0.205 | ahcy | -0.088 |
| stmn1b | 0.204 | otx5 | -0.084 |
| pou4f1 | 0.197 | hes2.2 | -0.084 |
| ywhag2 | 0.195 | ccna2 | -0.083 |
| stxbp1a | 0.195 | ndrg1b | -0.083 |
| zgc:65894 | 0.194 | dek | -0.081 |
| tubb5 | 0.193 | lbr | -0.080 |
| gng3 | 0.192 | mki67 | -0.080 |
| rbfox1 | 0.192 | msi1 | -0.080 |
| maptb | 0.189 | hes6 | -0.079 |
| zgc:153426 | 0.189 | mibp | -0.077 |
| syt2a | 0.188 | ddah2 | -0.076 |
| ebf3a | 0.188 | CABZ01005379.1 | -0.076 |
| pbx1a | 0.184 | dlgap5 | -0.075 |
| gap43 | 0.184 | zgc:110425 | -0.074 |
| eno2 | 0.184 | stmn1a | -0.073 |
| xpr1a | 0.183 | prdm1b | -0.073 |
| stx1b | 0.182 | XLOC-042899 | -0.073 |
| nrn1a | 0.182 | fbxo5 | -0.073 |
| isl2b | 0.181 | pcna | -0.072 |
| rbpms2b | 0.181 | rrm2 | -0.072 |
| dpysl3 | 0.181 | si:dkey-261m9.17 | -0.072 |
| ppp1r14ba | 0.180 | foxn4 | -0.072 |
| myt1la | 0.177 | zgc:110216 | -0.072 |
| id4 | 0.177 | XLOC-014477 | -0.071 |
| rtn1a | 0.176 | vsx1 | -0.071 |
| syt1a | 0.175 | tuba1b | -0.070 |
| cd99l2 | 0.174 | hist1h4l | -0.069 |