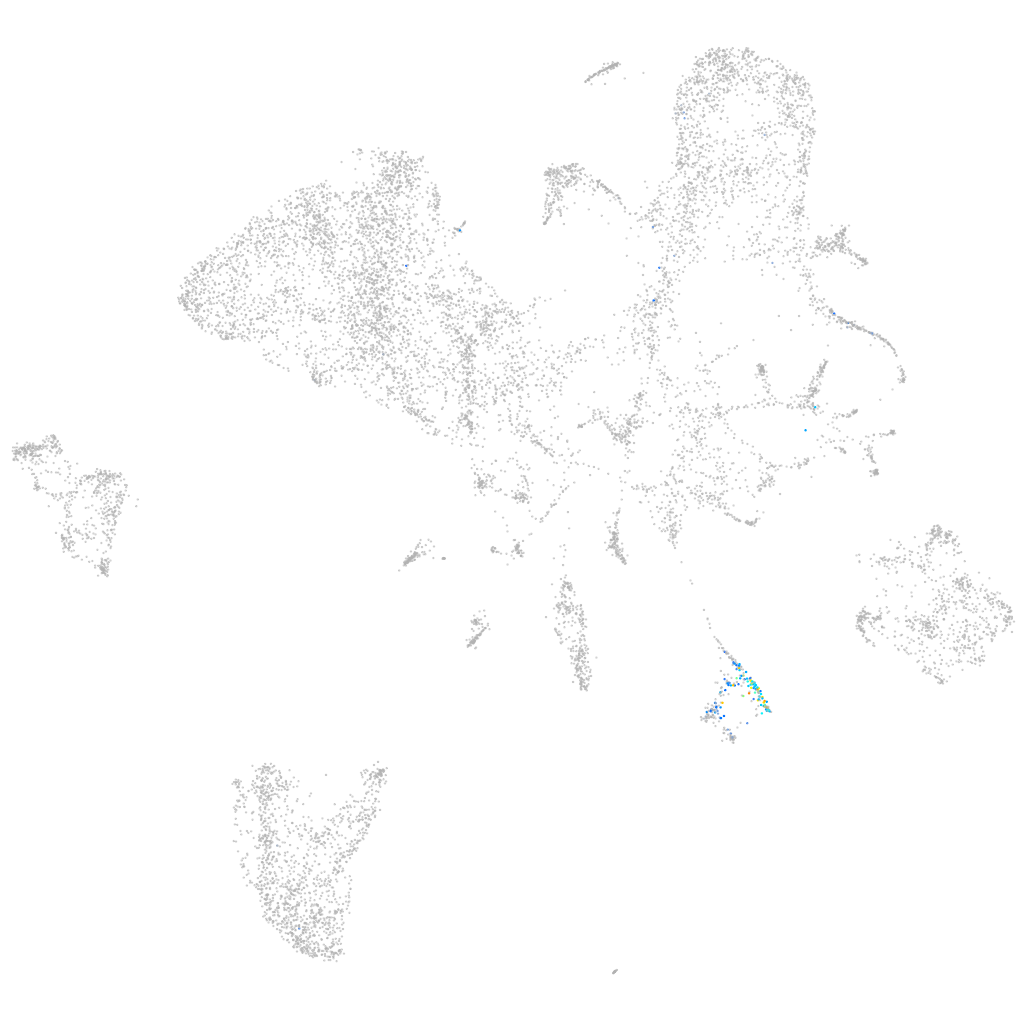
tektin 3
ZFIN 
Other cell groups


all cells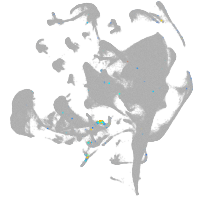 |
blastomeres |
PGCs |
endoderm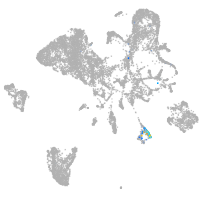 |
axial mesoderm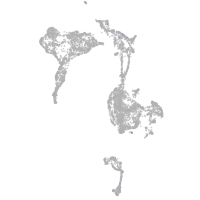 |
muscle 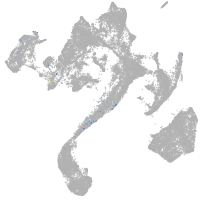 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 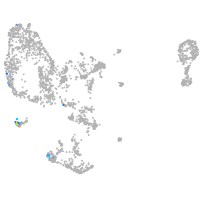 |
neural |
spinal cord / glia |
eye |
taste / olfactory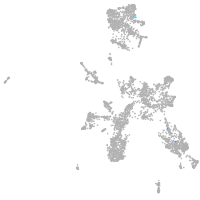 |
otic / lateral line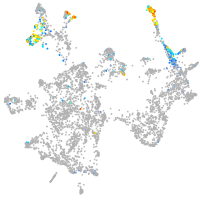 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmp22b | 0.566 | gapdh | -0.098 |
| CABZ01055365.1 | 0.564 | hspe1 | -0.097 |
| sim1b | 0.543 | gatm | -0.087 |
| cabp2b | 0.534 | gpx4a | -0.082 |
| mxra8b | 0.518 | fbp1b | -0.082 |
| b3gnt5a | 0.512 | aldob | -0.080 |
| lrata | 0.508 | bhmt | -0.080 |
| ctgfa | 0.499 | mdh1aa | -0.080 |
| arhgap29b | 0.491 | pklr | -0.079 |
| pnp4a | 0.488 | ahcy | -0.079 |
| spaca4l | 0.479 | gamt | -0.077 |
| spns2 | 0.476 | mat1a | -0.076 |
| FP102018.1 | 0.465 | si:dkey-16p21.8 | -0.076 |
| abca12 | 0.462 | apoc2 | -0.075 |
| spock3 | 0.459 | apoa4b.1 | -0.075 |
| sgms1 | 0.449 | lgals2b | -0.074 |
| cav1 | 0.441 | aqp12 | -0.074 |
| anxa3b | 0.435 | afp4 | -0.074 |
| f3a | 0.430 | npm1a | -0.074 |
| sim2 | 0.428 | apoa1b | -0.073 |
| si:dkey-30c15.2 | 0.421 | gcshb | -0.073 |
| cdc42ep1b | 0.416 | atp5mc3b | -0.071 |
| sftpba | 0.412 | gpd1b | -0.071 |
| cygb1 | 0.411 | apoc1 | -0.070 |
| cd151 | 0.410 | hspd1 | -0.068 |
| CU984579.1 | 0.407 | aldh6a1 | -0.068 |
| tgfb3 | 0.406 | abat | -0.068 |
| col4a6 | 0.405 | adh5 | -0.068 |
| col4a5 | 0.403 | ppifb | -0.066 |
| gna15.1 | 0.397 | grhprb | -0.066 |
| vwa1 | 0.395 | slc37a4a | -0.066 |
| rab32a | 0.391 | sult2st2 | -0.065 |
| zgc:63831 | 0.391 | clic5b | -0.065 |
| sim2.1 | 0.390 | slc16a10 | -0.065 |
| lama5 | 0.389 | g6pca.2 | -0.065 |