TEA domain family member 1a
ZFIN 
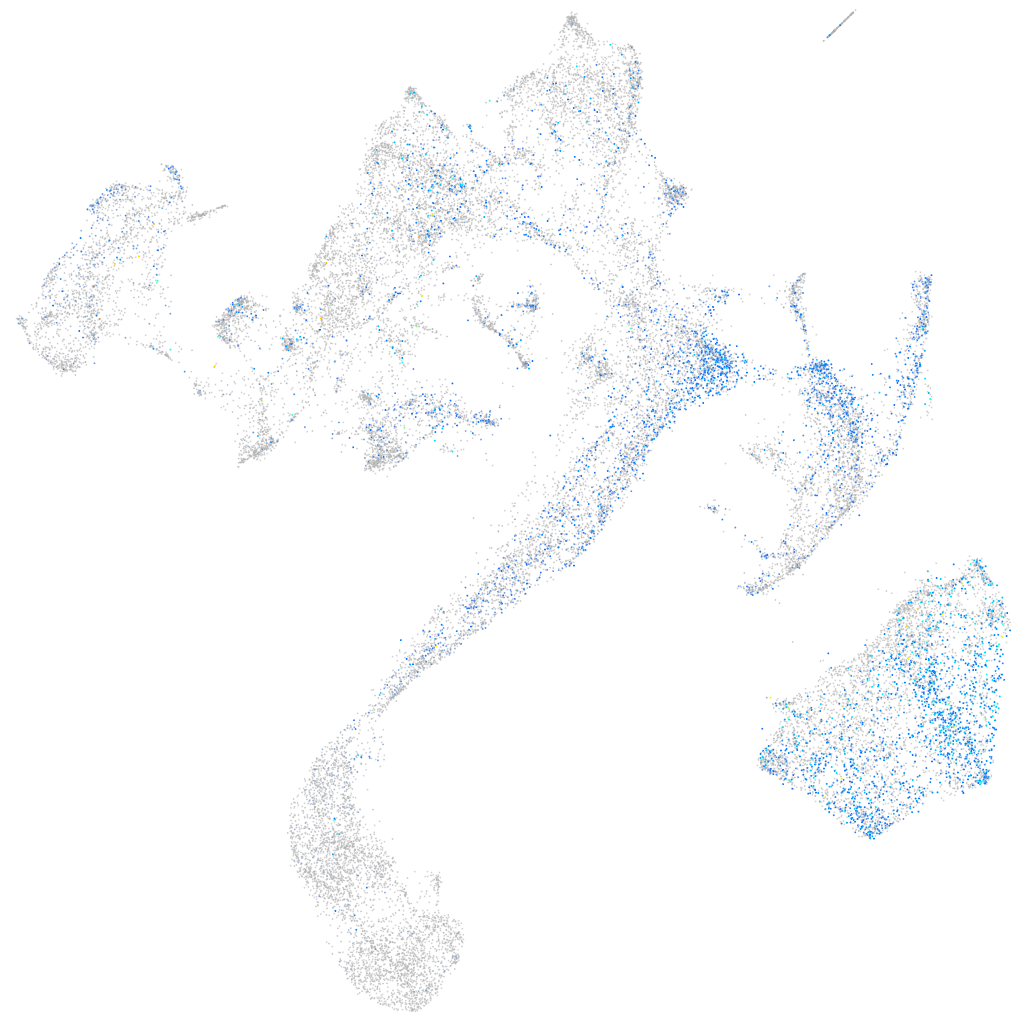
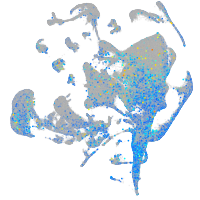
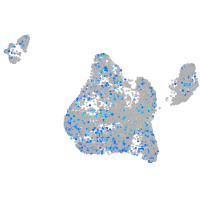
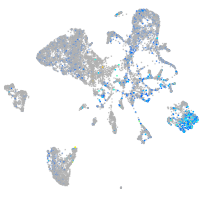
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpub | 0.258 | rpl37 | -0.227 |
| stm | 0.257 | zgc:114188 | -0.212 |
| anp32e | 0.256 | rps10 | -0.205 |
| hnrnpa1b | 0.250 | rps17 | -0.196 |
| efnb2b | 0.248 | fabp3 | -0.178 |
| ilf3b | 0.247 | actc1b | -0.176 |
| rbm4.3 | 0.247 | bhmt | -0.171 |
| NC-002333.4 | 0.247 | pvalb1 | -0.167 |
| nucks1a | 0.247 | idh2 | -0.167 |
| ncl | 0.242 | pvalb2 | -0.165 |
| acin1b | 0.242 | ckmb | -0.163 |
| srsf1a | 0.242 | ckma | -0.163 |
| syncrip | 0.242 | eef1da | -0.162 |
| hnrnpm | 0.240 | ak1 | -0.158 |
| hnrnpa1a | 0.240 | eef1b2 | -0.156 |
| ppig | 0.239 | mylpfa | -0.155 |
| srrm1 | 0.239 | mylz3 | -0.153 |
| acin1a | 0.239 | atp2a1 | -0.153 |
| fbl | 0.238 | eif4a1b | -0.152 |
| marcksb | 0.235 | atp5meb | -0.151 |
| hmga1a | 0.233 | tnnt3b | -0.150 |
| top1l | 0.233 | si:ch73-367p23.2 | -0.148 |
| im:7138239 | 0.233 | tnnc2 | -0.147 |
| apoeb | 0.233 | vdac3 | -0.146 |
| bms1 | 0.232 | tnni2a.4 | -0.145 |
| brd3a | 0.231 | tpi1b | -0.144 |
| nop58 | 0.231 | mdh2 | -0.144 |
| akap12b | 0.231 | eno3 | -0.144 |
| sp5l | 0.230 | ldb3b | -0.143 |
| safb | 0.229 | rps29 | -0.143 |
| nop56 | 0.228 | atp5f1b | -0.143 |
| dkc1 | 0.227 | pabpc4 | -0.142 |
| srrm2 | 0.227 | tnnt3a | -0.142 |
| cx43.4 | 0.226 | neb | -0.142 |
| hspb1 | 0.225 | eno1a | -0.142 |