tyrosyl-DNA phosphodiesterase 2a
ZFIN 
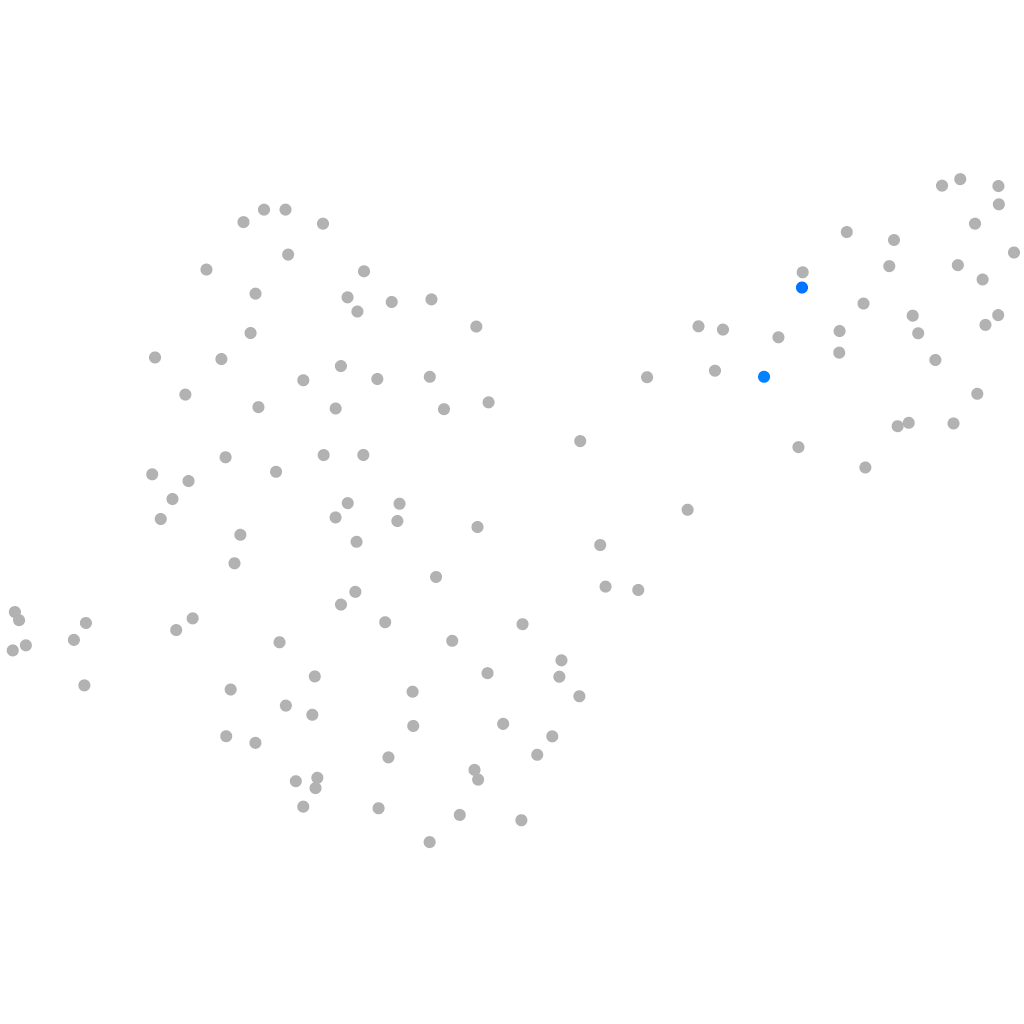
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcp4a | 0.844 | brd3a | -0.215 |
| gcnt4a | 0.818 | ybx1 | -0.201 |
| tgfb2 | 0.812 | nop56 | -0.198 |
| mapk8ip1a | 0.792 | hnrnpaba | -0.190 |
| BX897691.1 | 0.736 | g3bp1 | -0.172 |
| BX936364.1 | 0.736 | nucks1a | -0.172 |
| camk1ga | 0.736 | spcs2 | -0.172 |
| chia.2 | 0.736 | hnrnpa0a | -0.167 |
| cntn3a.1 | 0.736 | marcksb | -0.163 |
| cntn3b | 0.736 | ppig | -0.160 |
| CR318673.1 | 0.736 | hmgb1a | -0.158 |
| CR848746.1 | 0.736 | snrpa | -0.155 |
| cryba2a | 0.736 | snrpc | -0.154 |
| crybb1 | 0.736 | srrm1 | -0.153 |
| ctgfa | 0.736 | gtf2f1 | -0.146 |
| gabrr1 | 0.736 | srfbp1 | -0.146 |
| guca1b | 0.736 | npm1a | -0.144 |
| hcn4 | 0.736 | dek | -0.144 |
| her13 | 0.736 | banf1 | -0.142 |
| jdp2a | 0.736 | csnk1a1 | -0.142 |
| LOC100151570 | 0.736 | rbm7 | -0.141 |
| LOC101884613 | 0.736 | ddx23 | -0.140 |
| LOC101884995 | 0.736 | rrp15 | -0.140 |
| LOC108179959 | 0.736 | zgc:77849 | -0.139 |
| LOC562053 | 0.736 | cfl1 | -0.138 |
| lrch2 | 0.736 | ppp1r12a | -0.137 |
| matn4 | 0.736 | exosc10 | -0.136 |
| mir203a | 0.736 | hspd1 | -0.135 |
| mxtx2 | 0.736 | hnrnpm | -0.135 |
| ngfra | 0.736 | ptpn11a | -0.135 |
| npy8br | 0.736 | tcea1 | -0.133 |
| parvab | 0.736 | atp1a1a.1 | -0.132 |
| pclaf | 0.736 | tpx2 | -0.132 |
| prf1.9 | 0.736 | ctnnb1 | -0.132 |
| ptpn20 | 0.736 | akap12b | -0.131 |