L-threonine dehydrogenase
ZFIN 
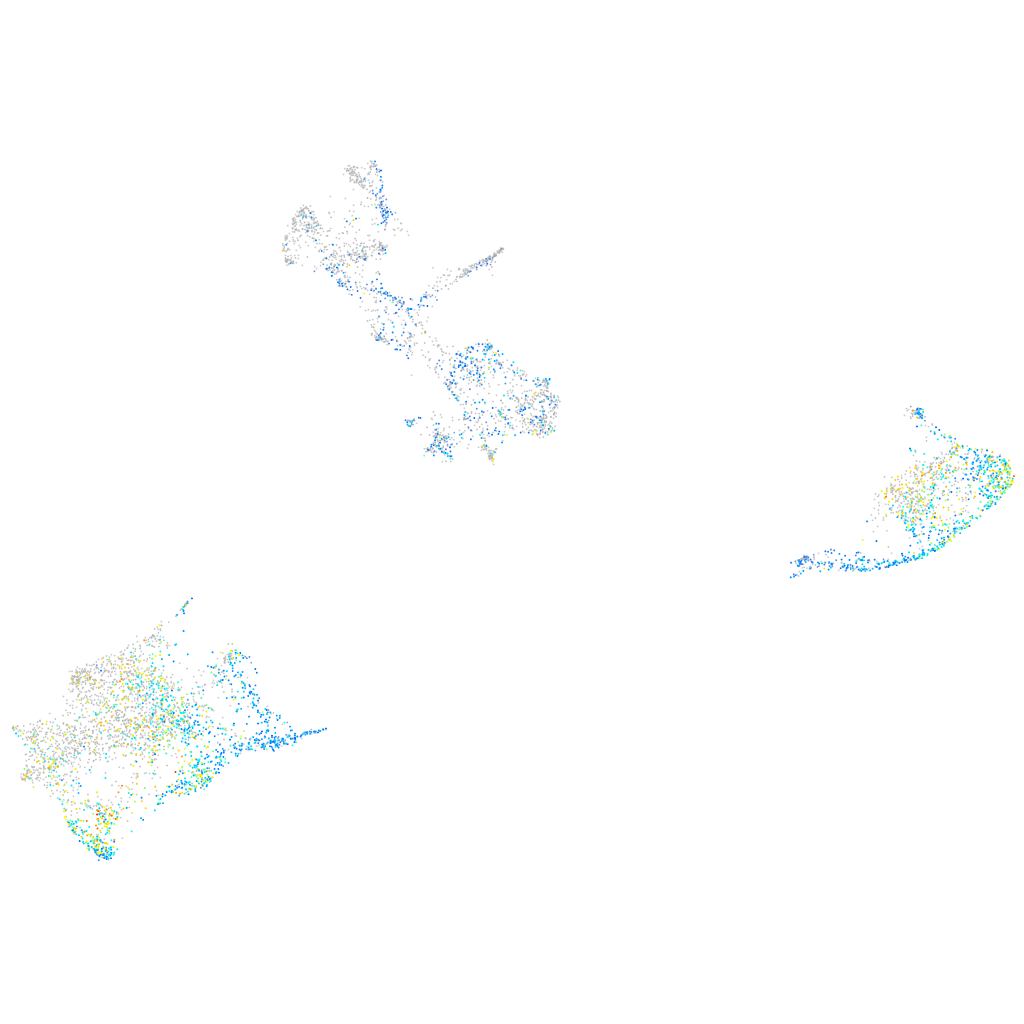
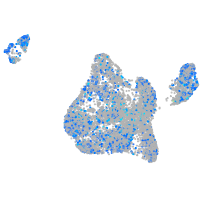
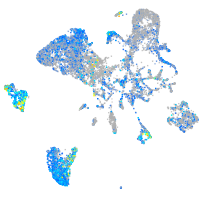
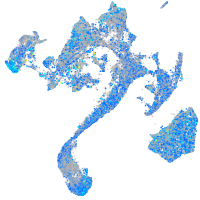
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| inka1b | 0.339 | defbl1 | -0.254 |
| rasd1 | 0.312 | apoda.1 | -0.253 |
| mitfa | 0.292 | gpnmb | -0.234 |
| cd63 | 0.286 | lypc | -0.232 |
| tfap2e | 0.286 | si:ch211-256m1.8 | -0.231 |
| si:zfos-943e10.1 | 0.283 | si:dkey-197i20.6 | -0.216 |
| tspan36 | 0.281 | mdkb | -0.207 |
| mlpha | 0.280 | akap12a | -0.207 |
| pah | 0.272 | si:ch73-89b15.3 | -0.200 |
| tmem243a | 0.271 | alx4a | -0.200 |
| qdpra | 0.258 | ptmaa | -0.196 |
| SPAG9 | 0.255 | unm-sa821 | -0.194 |
| pim1 | 0.254 | s100v2 | -0.193 |
| tspan10 | 0.254 | guk1a | -0.192 |
| bace2 | 0.251 | ponzr1 | -0.188 |
| id3 | 0.251 | sytl2b | -0.182 |
| opn5 | 0.249 | prx | -0.180 |
| LOC103910009 | 0.248 | alx4b | -0.179 |
| kcnj13 | 0.248 | ltk | -0.172 |
| pcbd1 | 0.246 | slc25a36a | -0.172 |
| xbp1 | 0.242 | cdh11 | -0.170 |
| slc3a2a | 0.241 | alx1 | -0.170 |
| cdh1 | 0.237 | CABZ01072077.1 | -0.163 |
| sik1 | 0.237 | tcirg1a | -0.157 |
| pcdh10a | 0.237 | si:ch211-105c13.3 | -0.156 |
| rab34b | 0.235 | psat1 | -0.156 |
| syngr1a | 0.235 | fhl3a | -0.154 |
| tyr | 0.235 | cx43 | -0.153 |
| rab38 | 0.235 | fhl2a | -0.151 |
| gstt1a | 0.232 | si:ch211-243a20.3 | -0.150 |
| si:ch211-195b13.1 | 0.231 | pltp | -0.150 |
| kita | 0.231 | sparc | -0.149 |
| C12orf75 | 0.231 | ptmab | -0.148 |
| ctsla | 0.228 | tuba1b | -0.148 |
| socs3a | 0.227 | CT737162.3 | -0.148 |