Tctex1 domain containing 2
ZFIN 
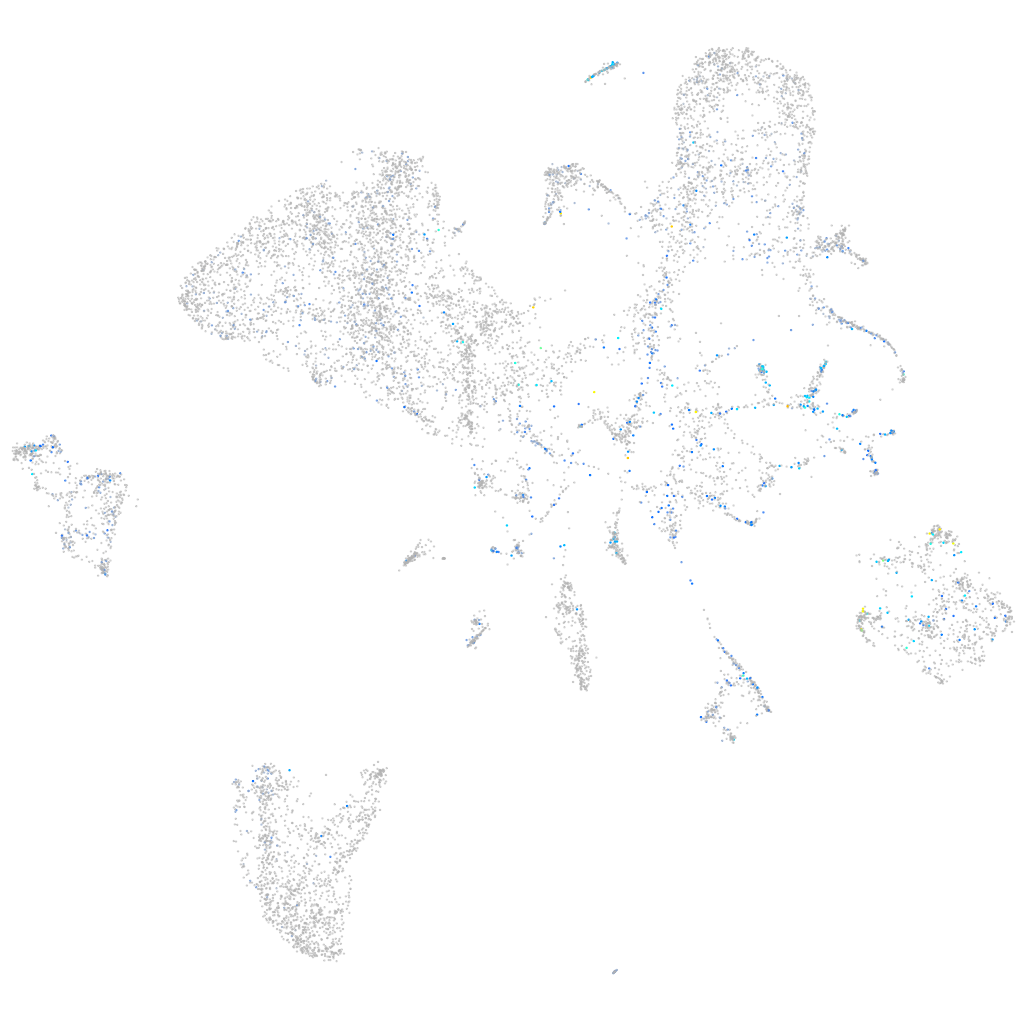
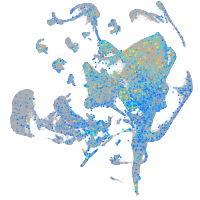
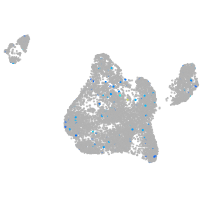
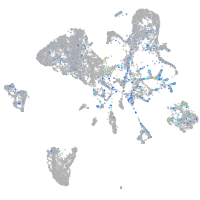
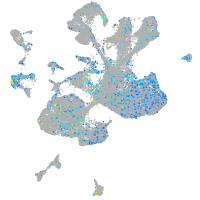
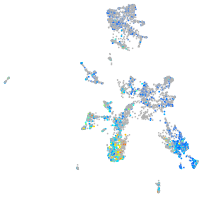
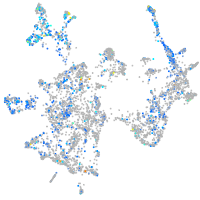
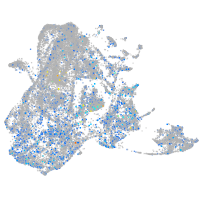
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.216 | gamt | -0.151 |
| pcsk1nl | 0.191 | gatm | -0.148 |
| zgc:101731 | 0.191 | gapdh | -0.143 |
| insm1b | 0.190 | bhmt | -0.132 |
| neurod1 | 0.184 | ahcy | -0.125 |
| fev | 0.176 | nupr1b | -0.119 |
| capsla | 0.171 | pnp4b | -0.116 |
| egr4 | 0.171 | mat1a | -0.115 |
| vat1 | 0.168 | apoa4b.1 | -0.115 |
| got1l1 | 0.168 | afp4 | -0.114 |
| slc35g2a | 0.167 | apoa1b | -0.113 |
| hsbp1a | 0.166 | aqp12 | -0.112 |
| pax4 | 0.163 | fbp1b | -0.111 |
| nkx2.2a | 0.156 | agxtb | -0.109 |
| hmgn6 | 0.156 | aldob | -0.108 |
| si:ch211-222l21.1 | 0.156 | apoc2 | -0.104 |
| gpc1a | 0.156 | apobb.1 | -0.102 |
| lysmd2 | 0.155 | scp2a | -0.101 |
| gapdhs | 0.155 | apoa2 | -0.101 |
| rasd4 | 0.155 | abat | -0.099 |
| TCIM (1 of many) | 0.154 | fabp10a | -0.096 |
| hepacam2 | 0.153 | tfa | -0.096 |
| si:ch1073-429i10.3.1 | 0.152 | glud1b | -0.096 |
| c2cd4a | 0.152 | cx32.3 | -0.095 |
| mir7a-1 | 0.151 | dap | -0.095 |
| h3f3d | 0.150 | gpx4a | -0.094 |
| h3f3c | 0.150 | ces2 | -0.094 |
| hmgn2 | 0.149 | serpina1 | -0.094 |
| scgn | 0.149 | slc27a2a | -0.093 |
| rprmb | 0.148 | fetub | -0.093 |
| ywhabl | 0.148 | hao1 | -0.093 |
| sumo3a | 0.148 | grhprb | -0.093 |
| oaz1b | 0.148 | apom | -0.093 |
| insm1a | 0.148 | LOC110437731 | -0.093 |
| si:dkey-153k10.9 | 0.147 | zgc:123103 | -0.093 |