thromboxane A2 receptor
ZFIN 
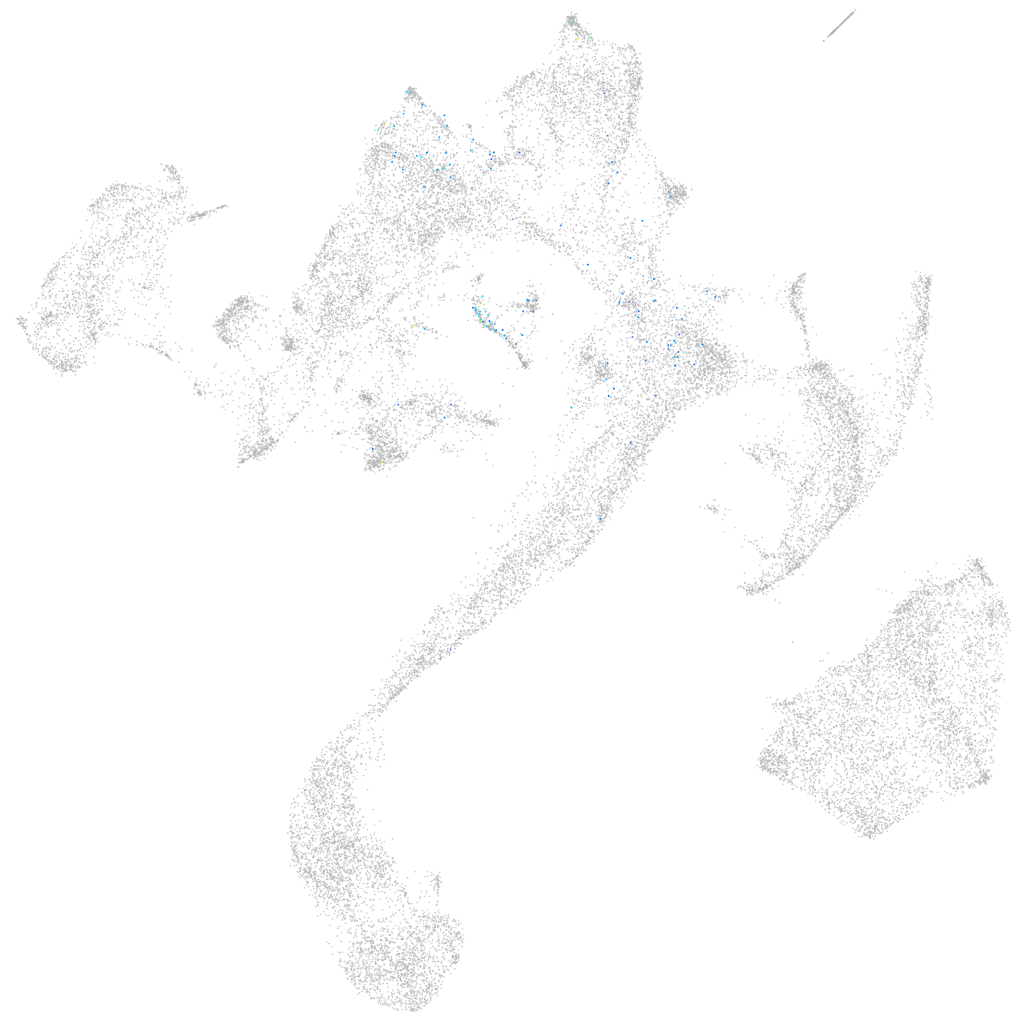
Other cell groups


all cells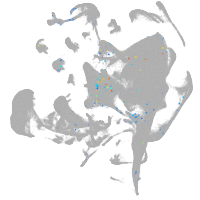 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 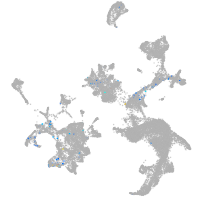 |
pronephros 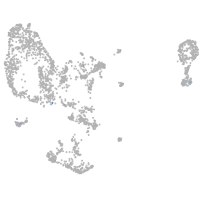 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pitx1 | 0.140 | cycsb | -0.046 |
| pimr80 | 0.138 | hspe1 | -0.041 |
| pcolcea | 0.123 | atp5f1e | -0.038 |
| ppp1r42 | 0.116 | aldoab | -0.037 |
| prokr1b | 0.112 | cox7b | -0.036 |
| FQ323119.1 | 0.110 | atp2a1 | -0.035 |
| LO018585.1 | 0.104 | tmem38a | -0.035 |
| cfd | 0.104 | gapdh | -0.034 |
| rhag | 0.104 | ckmb | -0.034 |
| CR925719.1 | 0.103 | mybphb | -0.034 |
| si:ch1073-138d10.2 | 0.100 | ckma | -0.034 |
| si:ch211-147m20.3 | 0.099 | ttn.2 | -0.033 |
| LOC101885014 | 0.098 | acta1a | -0.033 |
| LOC110439867 | 0.095 | ttn.1 | -0.033 |
| col6a3 | 0.092 | desma | -0.033 |
| ftr82 | 0.092 | ldb3a | -0.033 |
| col7a1l | 0.091 | ak1 | -0.033 |
| tmem88b | 0.089 | cox6c | -0.032 |
| entpd3 | 0.088 | neb | -0.032 |
| klf6a | 0.087 | cdx4 | -0.032 |
| col6a2 | 0.087 | srl | -0.032 |
| col6a1 | 0.086 | ldb3b | -0.032 |
| twist1a | 0.086 | uqcrh | -0.032 |
| lum | 0.085 | actc1b | -0.032 |
| agrp | 0.085 | cav3 | -0.031 |
| CABZ01027646.1 | 0.085 | hspb1 | -0.031 |
| si:ch211-92l17.1 | 0.083 | fxr2 | -0.031 |
| alx4b | 0.083 | klhl41b | -0.031 |
| cavin2b | 0.082 | actc1a | -0.031 |
| si:ch1073-459j12.1 | 0.082 | tnnc2 | -0.031 |
| tril | 0.081 | chchd10 | -0.030 |
| FRMD1 | 0.081 | pabpc4 | -0.030 |
| s100v2 | 0.080 | zgc:101853 | -0.030 |
| aoc1 | 0.080 | txlnbb | -0.030 |
| mxra8b | 0.079 | ndufab1b | -0.030 |