TatD DNase domain containing 2
ZFIN 
Other cell groups


all cells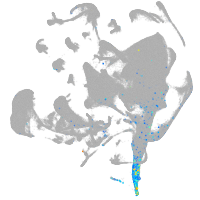 |
blastomeres |
PGCs |
endoderm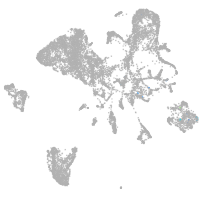 |
axial mesoderm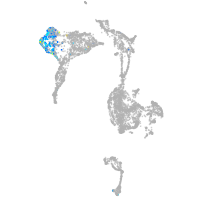 |
muscle 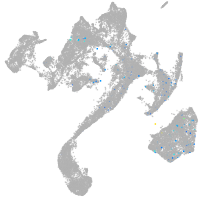 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-001602 | 0.115 | ckbb | -0.031 |
| si:dkeyp-32g11.8 | 0.112 | gpm6aa | -0.029 |
| siglec15l | 0.098 | CR383676.1 | -0.028 |
| CR792441.1 | 0.080 | pvalb1 | -0.028 |
| si:dkey-30j22.1 | 0.079 | nova2 | -0.027 |
| si:ch1073-166e24.4 | 0.078 | si:dkeyp-75h12.5 | -0.026 |
| CABZ01074470.1 | 0.075 | rtn1a | -0.026 |
| LOC103909808 | 0.072 | actc1b | -0.026 |
| CR382363.1 | 0.066 | pvalb2 | -0.025 |
| fgf9 | 0.066 | marcksl1b | -0.024 |
| kcnip2 | 0.062 | myt1b | -0.023 |
| LOC796940 | 0.059 | gpm6ab | -0.023 |
| CR926130.1 | 0.058 | tmsb | -0.022 |
| BX004840.2 | 0.057 | ube2d4 | -0.022 |
| npm1a | 0.053 | CU634008.1 | -0.022 |
| cdx4 | 0.053 | COX3 | -0.021 |
| hspb1 | 0.052 | CU467822.1 | -0.021 |
| nhp2 | 0.050 | elavl3 | -0.021 |
| znfl2a | 0.048 | mylpfa | -0.021 |
| dut | 0.048 | gpm6bb | -0.021 |
| CT027744.2 | 0.048 | elavl4 | -0.021 |
| aqp9b | 0.048 | hbbe1.3 | -0.020 |
| rpa2 | 0.047 | gatad2b | -0.020 |
| apela | 0.047 | nova1 | -0.020 |
| sp5l | 0.047 | slc1a2b | -0.020 |
| nop10 | 0.046 | gng3 | -0.019 |
| anp32b | 0.046 | krt4 | -0.019 |
| mttp | 0.046 | stmn1b | -0.019 |
| LOC110439365 | 0.045 | vamp2 | -0.019 |
| rfc5 | 0.045 | dbn1 | -0.019 |
| BX927258.1 | 0.045 | hbae3 | -0.019 |
| homer3a | 0.045 | slc6a1b | -0.019 |
| mcm7 | 0.045 | si:dkey-276j7.1 | -0.019 |
| LOC103911653 | 0.044 | atp6v0cb | -0.019 |
| dkc1 | 0.044 | atp1b4 | -0.018 |




