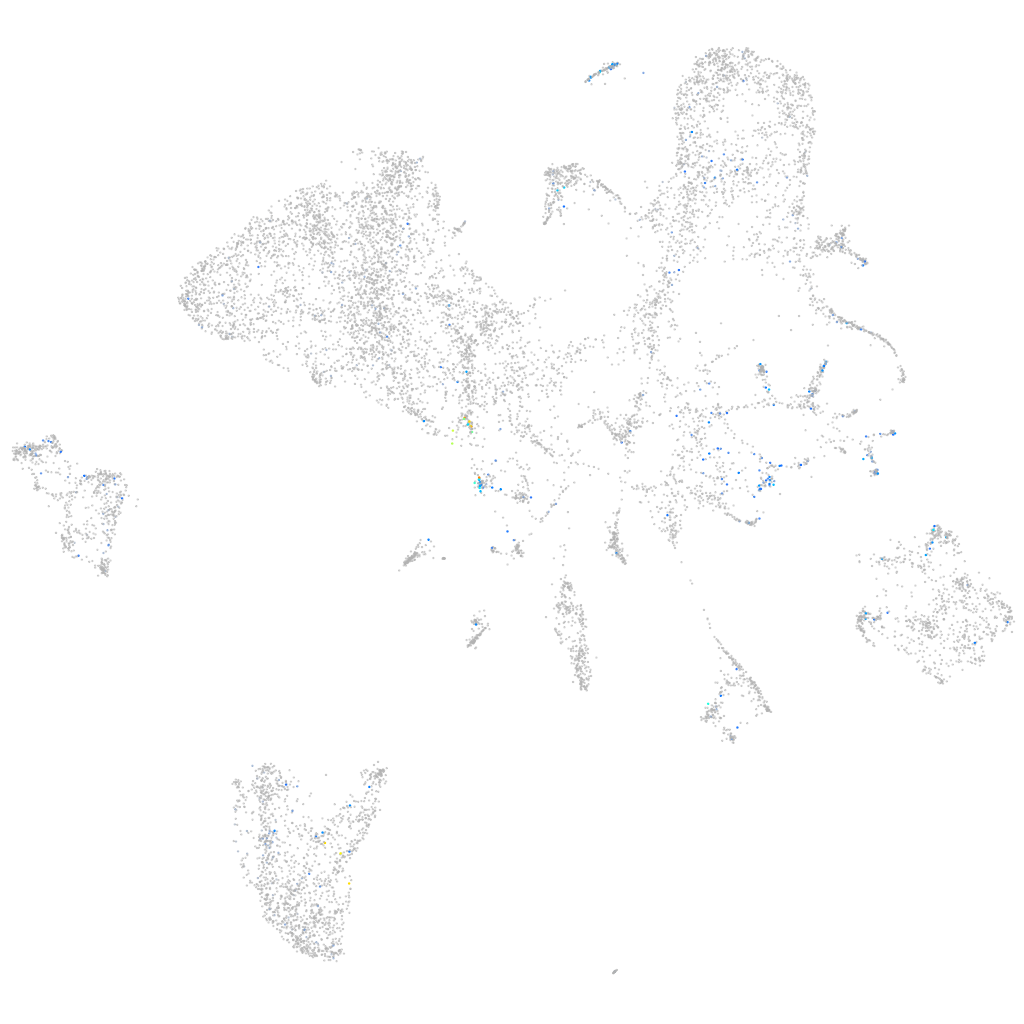
synaptophysin b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gabrg2 | 0.328 | aldob | -0.098 |
| syt5b | 0.280 | ahcy | -0.097 |
| cntn1b | 0.273 | gamt | -0.097 |
| lactbl1a | 0.273 | gapdh | -0.084 |
| vsx1 | 0.268 | gatm | -0.084 |
| nrn1lb | 0.251 | gstt1a | -0.082 |
| snap25b | 0.249 | fbp1b | -0.080 |
| si:ch211-237i5.4 | 0.235 | aldh6a1 | -0.080 |
| lrrc24 | 0.230 | apoa4b.1 | -0.080 |
| sncgb | 0.221 | scp2a | -0.074 |
| vamp1 | 0.218 | gstr | -0.074 |
| atp6v0cb | 0.217 | apoc2 | -0.073 |
| calb2b | 0.213 | rgrb | -0.072 |
| LOC108183447 | 0.212 | pnp4b | -0.072 |
| cabp2a | 0.212 | gpx4a | -0.071 |
| sv2a | 0.207 | eno3 | -0.070 |
| vsnl1b | 0.203 | afp4 | -0.070 |
| LOC110438143 | 0.203 | nipsnap3a | -0.070 |
| CABZ01038709.1 | 0.203 | agxta | -0.069 |
| dkk3a | 0.203 | mat1a | -0.069 |
| cacna1fb | 0.202 | bhmt | -0.069 |
| gnat1 | 0.202 | eef1da | -0.069 |
| KIRREL3 | 0.201 | pgm1 | -0.068 |
| fxyd6 | 0.201 | cox7a1 | -0.068 |
| si:ch1073-83n3.2 | 0.198 | suclg1 | -0.068 |
| tex2l | 0.197 | isoc2 | -0.067 |
| kcnv2a | 0.194 | aldh1l1 | -0.067 |
| rs1a | 0.194 | cfhl4 | -0.067 |
| lhfpl3 | 0.191 | cx32.3 | -0.067 |
| gng3 | 0.190 | hspe1 | -0.067 |
| dscaml1 | 0.189 | fetub | -0.067 |
| BX663503.1 | 0.189 | dap | -0.067 |
| cplx4a | 0.187 | serpinf2b | -0.067 |
| tdh2 | 0.187 | apobb.1 | -0.067 |
| atp6v0a1b | 0.186 | gstz1 | -0.067 |