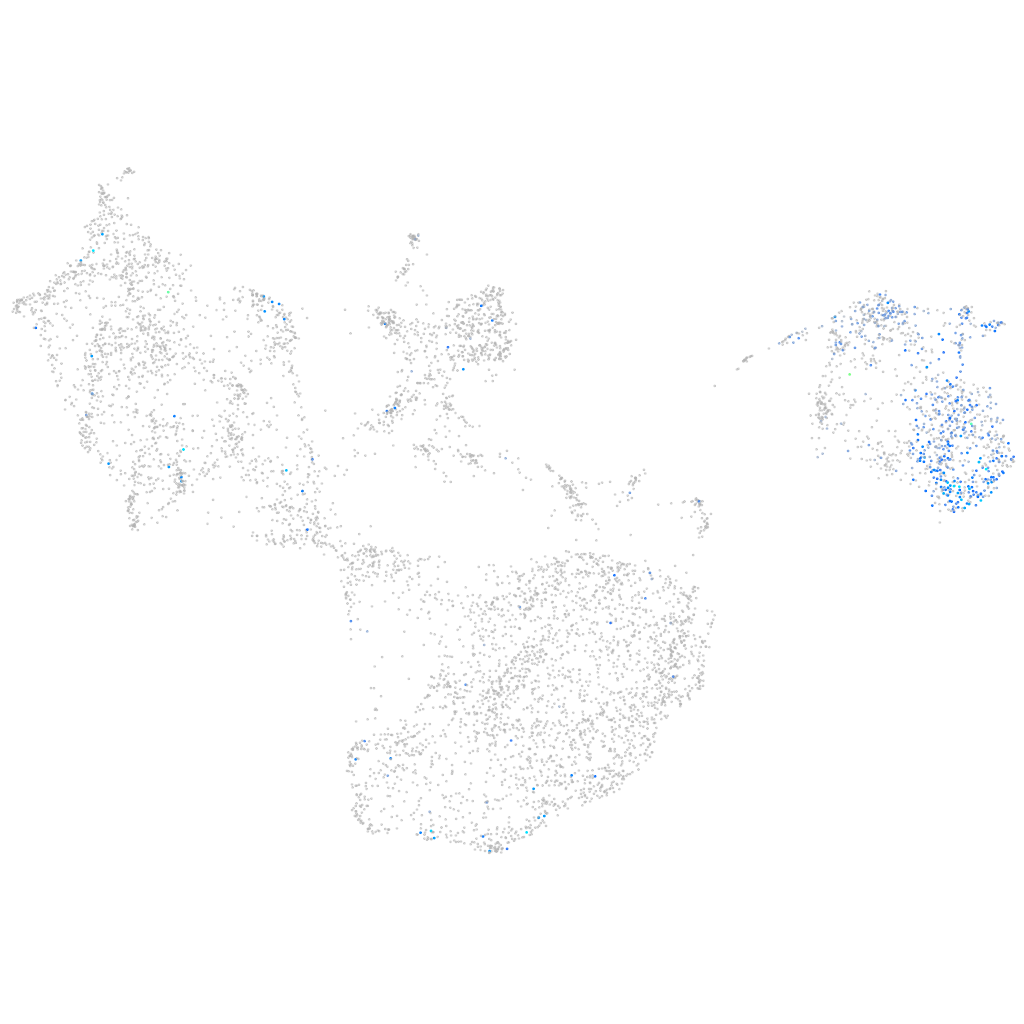
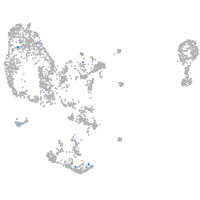
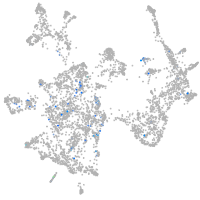
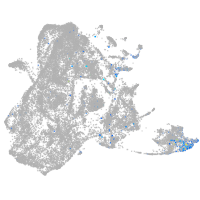
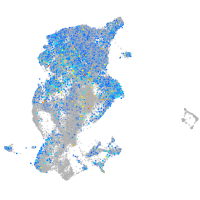
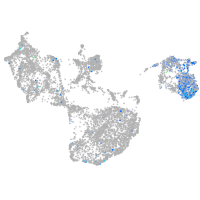
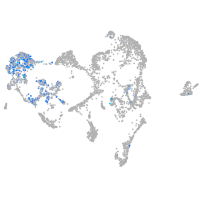
switching B cell complex subunit SWAP70a
ZFIN 
Expression by stage/cluster


Correlated gene expression