"sulfotransferase family 1, cytosolic sulfotransferase 5"
ZFIN 
Other cell groups


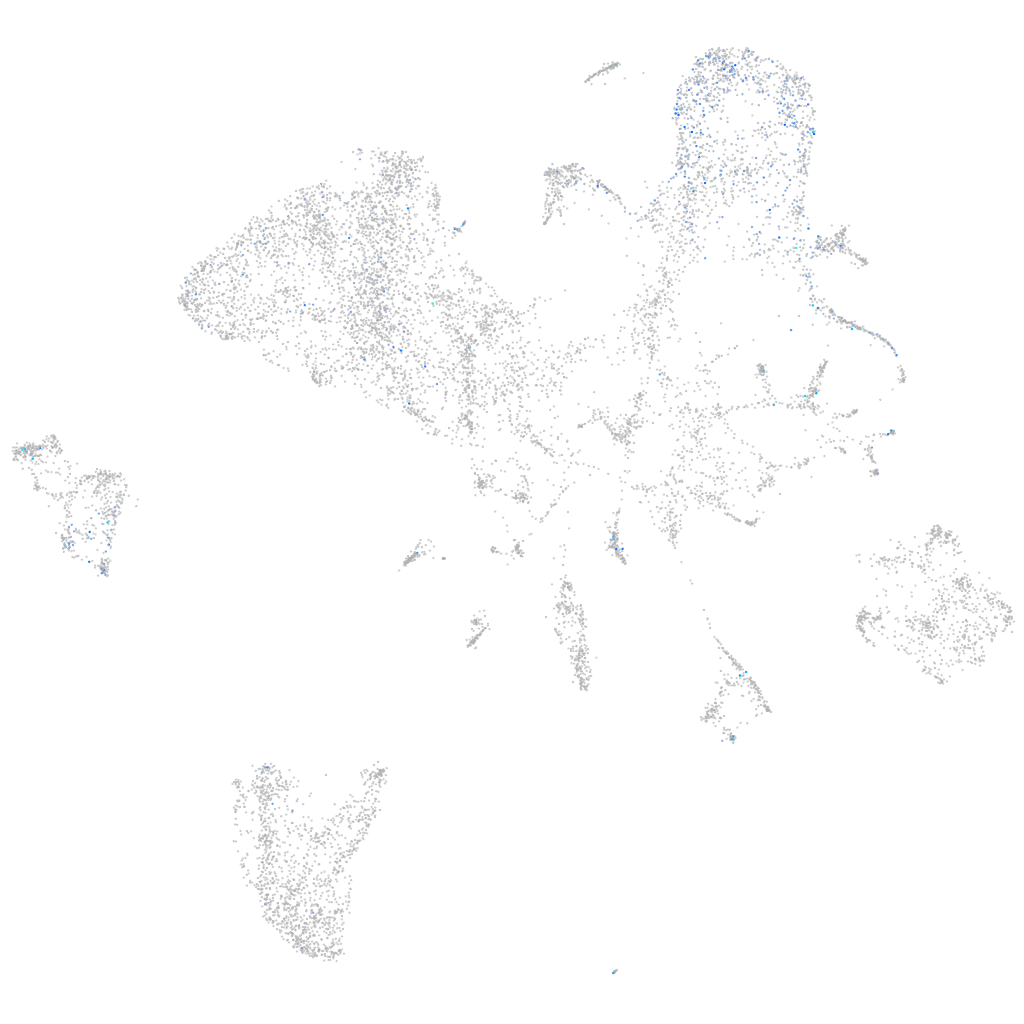
all cells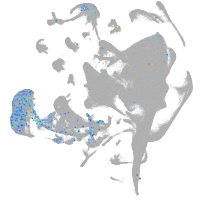 |
blastomeres |
PGCs |
endoderm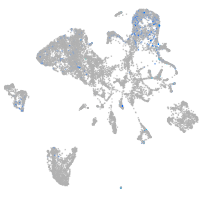 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 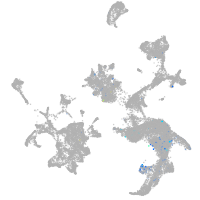 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:153968 | 0.200 | aqp12 | -0.093 |
| ace2 | 0.199 | bhmt | -0.086 |
| si:dkey-36i7.3 | 0.196 | fabp3 | -0.084 |
| si:ch211-133l5.7 | 0.195 | marcksb | -0.071 |
| atp1a1a.4 | 0.195 | apoc1 | -0.069 |
| AL831745.1 | 0.195 | cd81a | -0.068 |
| cd36 | 0.194 | cx43.4 | -0.065 |
| zgc:158846 | 0.194 | rtn1a | -0.065 |
| krt97 | 0.193 | marcksl1b | -0.062 |
| dhrs1 | 0.192 | hspb1 | -0.061 |
| zgc:172079 | 0.192 | zgc:136461 | -0.061 |
| mogat2 | 0.189 | ela2l | -0.060 |
| sult6b1 | 0.189 | cpb1 | -0.060 |
| aoc1 | 0.188 | cel.1 | -0.059 |
| aqp8a.2 | 0.188 | c6ast4 | -0.059 |
| vil1 | 0.187 | prss59.1 | -0.059 |
| plac8.1 | 0.187 | ctrl | -0.059 |
| chia.2 | 0.186 | prss1 | -0.059 |
| cyp2x9 | 0.186 | agxtb | -0.059 |
| krt92 | 0.185 | zgc:112160 | -0.059 |
| slc15a1b | 0.184 | sycn.2 | -0.058 |
| cyp3c3 | 0.183 | ela3l | -0.058 |
| ada | 0.183 | cel.2 | -0.058 |
| anxa2b | 0.183 | fep15 | -0.058 |
| zanl | 0.183 | amy2a | -0.058 |
| slc6a19a.2 | 0.183 | si:ch211-240l19.5 | -0.057 |
| si:ch211-161h7.8 | 0.183 | cpa4 | -0.057 |
| pdzk1 | 0.182 | cpn1 | -0.057 |
| cdh17 | 0.182 | zgc:92137 | -0.057 |
| gsto2 | 0.182 | ela2 | -0.057 |
| cpo | 0.182 | ppdpfa | -0.057 |
| meltf | 0.182 | cpa5 | -0.056 |
| rtn4b | 0.181 | prss59.2 | -0.056 |
| mansc1 | 0.180 | pdia2 | -0.056 |
| ethe1 | 0.180 | eif4ebp3 | -0.056 |