"sulfotransferase family 1, cytosolic sulfotransferase 1"
ZFIN 
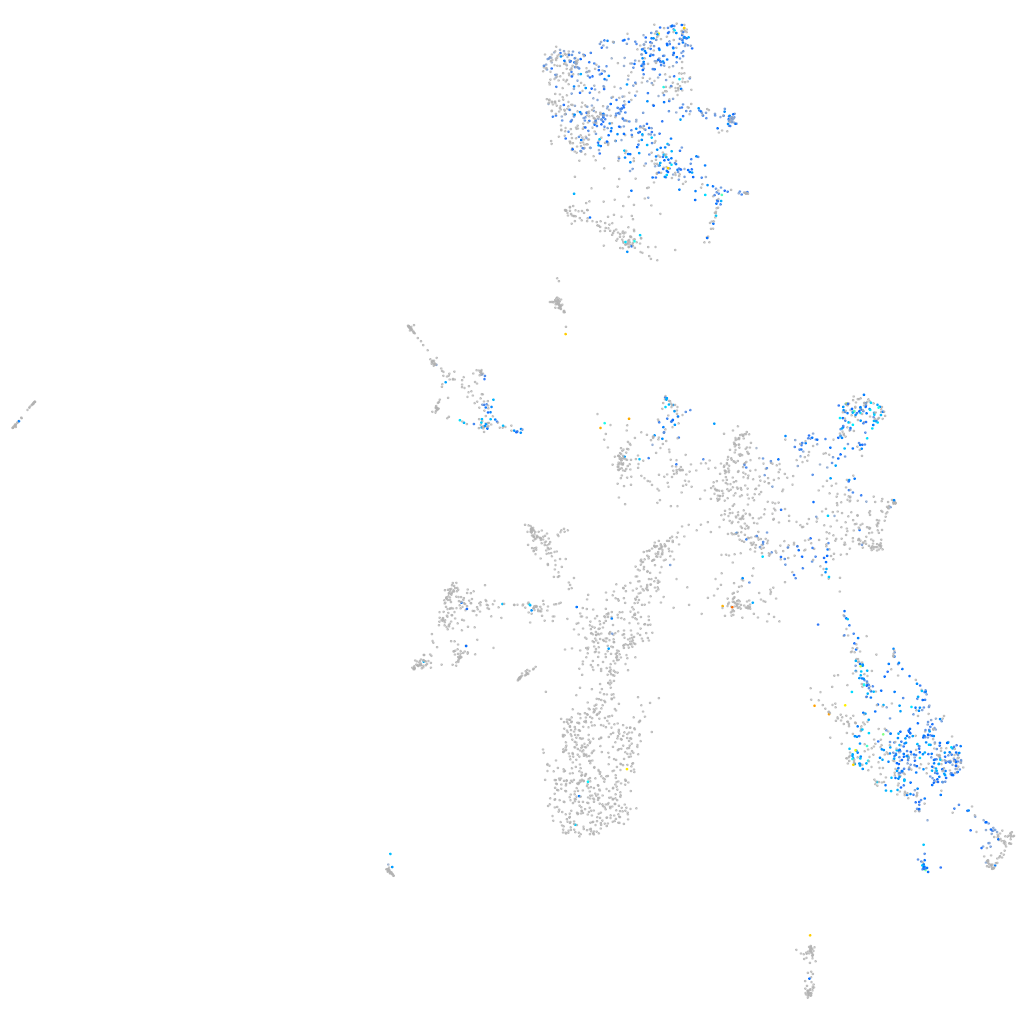
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aqp3a | 0.469 | elavl3 | -0.243 |
| zgc:158343 | 0.428 | insm1a | -0.205 |
| pycard | 0.425 | rtn1a | -0.196 |
| cfl1l | 0.411 | tuba1a | -0.193 |
| sparc | 0.404 | tmeff1b | -0.193 |
| krt4 | 0.400 | stmn1b | -0.187 |
| s100a10b | 0.393 | ebf3a | -0.180 |
| zgc:162730 | 0.379 | epb41a | -0.180 |
| cldne | 0.373 | rnasekb | -0.175 |
| s100v1 | 0.372 | atp6v0cb | -0.171 |
| krt8 | 0.371 | hmgb3a | -0.165 |
| her9 | 0.365 | NC-002333.4 | -0.165 |
| lgals3b | 0.365 | si:dkey-56m19.5 | -0.165 |
| krt18a.1 | 0.364 | CU467822.1 | -0.165 |
| cebpd | 0.360 | myt1a | -0.163 |
| tmsb1 | 0.358 | sncb | -0.161 |
| gsta.1 | 0.357 | sox11a | -0.160 |
| capns1a | 0.355 | nova2 | -0.158 |
| cnn2 | 0.353 | celf3a | -0.156 |
| dnase1l4.1 | 0.353 | zgc:65894 | -0.156 |
| tmem176l.4 | 0.347 | si:dkey-276j7.1 | -0.155 |
| krt18b | 0.343 | si:dkeyp-75h12.5 | -0.155 |
| malb | 0.341 | insm1b | -0.154 |
| si:dkey-16l2.20 | 0.340 | gng3 | -0.151 |
| krt1-c5 | 0.339 | tspan2a | -0.151 |
| sdc4 | 0.337 | ptmab | -0.147 |
| sult6b1 | 0.336 | cnrip1a | -0.145 |
| cebpb | 0.336 | gnao1a | -0.145 |
| elf3 | 0.336 | neurod1 | -0.144 |
| eef1da | 0.336 | elavl4 | -0.143 |
| tagln2 | 0.331 | LOC100537342 | -0.142 |
| krt97 | 0.326 | uncx | -0.141 |
| actb2 | 0.326 | atp2b1b | -0.140 |
| ahnak | 0.325 | ebf2 | -0.140 |
| her6 | 0.322 | tuba1c | -0.140 |