"succinate-CoA ligase, alpha subunit"
ZFIN 
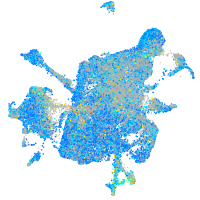
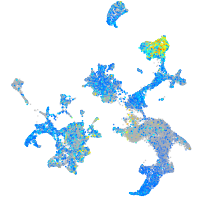
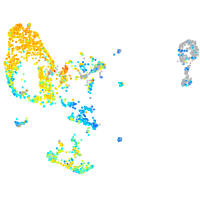
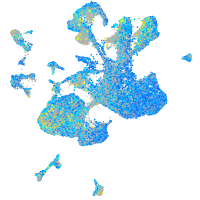
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldoab | 0.648 | ptmab | -0.597 |
| srl | 0.630 | si:ch73-1a9.3 | -0.586 |
| tmem38a | 0.619 | si:ch211-222l21.1 | -0.553 |
| actc1b | 0.619 | pabpc1a | -0.545 |
| desma | 0.612 | hmgb2a | -0.516 |
| acta1a | 0.611 | hmgb2b | -0.511 |
| atp5if1b | 0.607 | hsp90ab1 | -0.509 |
| atp5f1b | 0.593 | anp32a | -0.508 |
| ttn.1 | 0.591 | cirbpa | -0.492 |
| ttn.2 | 0.591 | seta | -0.482 |
| ak1 | 0.591 | si:ch73-281n10.2 | -0.479 |
| atp5mc1 | 0.588 | nucks1a | -0.475 |
| atp5meb | 0.588 | acin1a | -0.472 |
| atp2a1 | 0.586 | cx43.4 | -0.467 |
| cox7a2a | 0.583 | anp32b | -0.463 |
| cav3 | 0.580 | si:ch211-288g17.3 | -0.463 |
| actc1a | 0.579 | hnrnpa1b | -0.463 |
| ckmb | 0.576 | hmgn7 | -0.458 |
| CABZ01072309.1 | 0.575 | hnrnpaba | -0.456 |
| gapdh | 0.573 | fthl27 | -0.455 |
| CABZ01078594.1 | 0.573 | khdrbs1a | -0.453 |
| casq2 | 0.573 | hmga1a | -0.453 |
| atp5mc3b | 0.570 | marcksb | -0.452 |
| cox17 | 0.565 | tuba8l4 | -0.451 |
| ckma | 0.564 | hmgn2 | -0.450 |
| eno3 | 0.555 | hnrnpa0b | -0.448 |
| tpi1b | 0.551 | marcksl1b | -0.445 |
| cox5aa | 0.549 | cirbpb | -0.442 |
| txlnbb | 0.549 | h3f3d | -0.440 |
| mdh2 | 0.548 | hnrnpa1a | -0.434 |
| neb | 0.545 | hnrnpub | -0.433 |
| mybphb | 0.545 | hnrnpabb | -0.429 |
| ldb3a | 0.544 | hmgb1b | -0.425 |
| pgam2 | 0.541 | top1l | -0.423 |
| vdac2 | 0.538 | syncrip | -0.422 |