"striatin, calmodulin binding protein 3"
ZFIN 
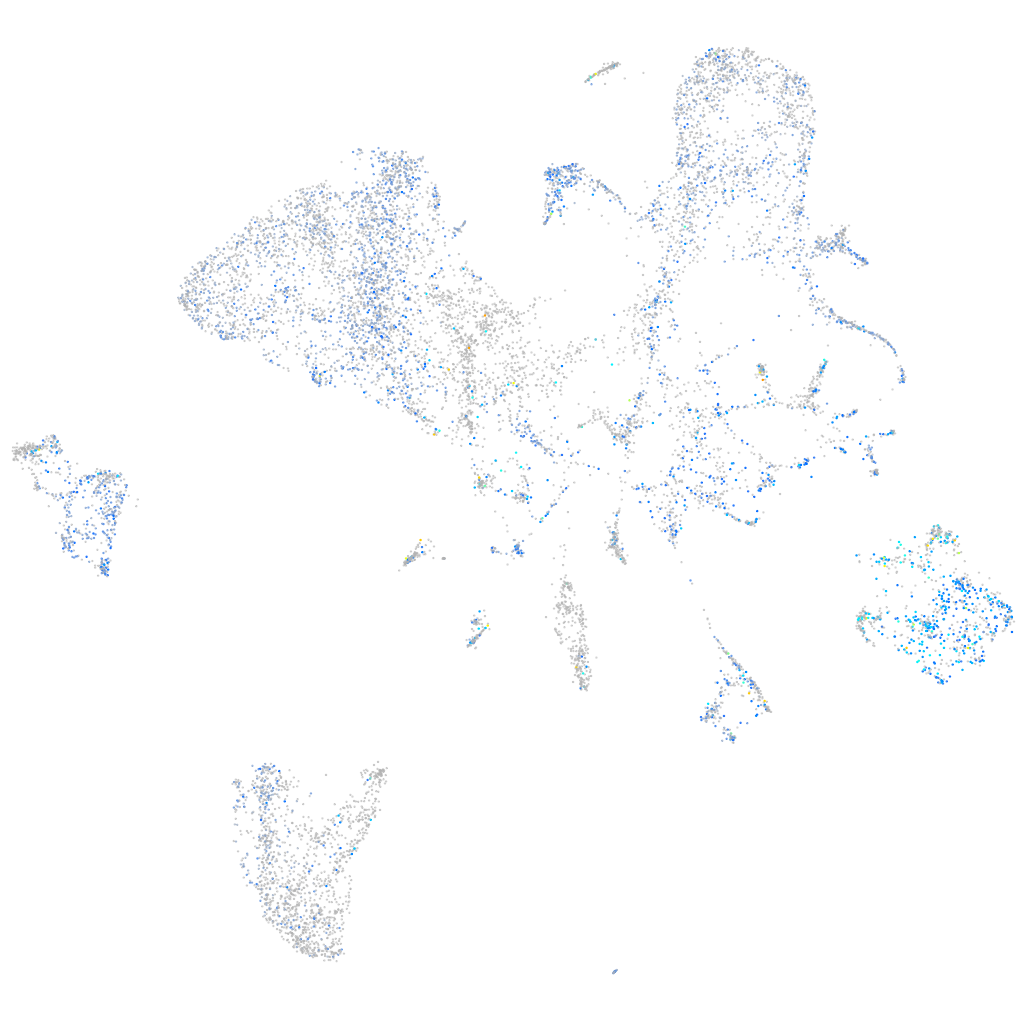
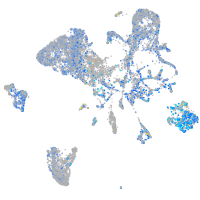
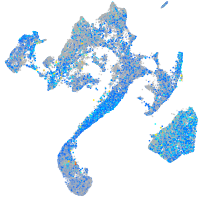
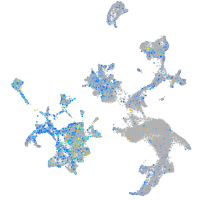
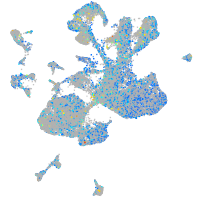
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| acin1a | 0.276 | rpl37 | -0.219 |
| ilf3b | 0.270 | rps10 | -0.201 |
| srrm2 | 0.268 | zgc:114188 | -0.192 |
| akap12b | 0.266 | gamt | -0.183 |
| hnrnpa1b | 0.266 | ahcy | -0.183 |
| nucks1a | 0.264 | zgc:92744 | -0.172 |
| hnrnpm | 0.262 | gapdh | -0.167 |
| si:ch211-152c2.3 | 0.262 | gatm | -0.161 |
| hnrnpa1a | 0.262 | rps17 | -0.160 |
| hspb1 | 0.261 | bhmt | -0.155 |
| asph | 0.261 | dap | -0.140 |
| smc1al | 0.260 | nme2b.1 | -0.139 |
| hnrnpub | 0.260 | aqp12 | -0.133 |
| marcksb | 0.257 | nupr1b | -0.131 |
| NC-002333.4 | 0.256 | prss1 | -0.124 |
| smarca4a | 0.255 | prss59.2 | -0.124 |
| ppig | 0.253 | suclg1 | -0.123 |
| anp32a | 0.253 | CELA1 (1 of many) | -0.123 |
| rbm4.3 | 0.252 | ctrb1 | -0.122 |
| top1l | 0.251 | eno3 | -0.121 |
| syncrip | 0.251 | gpx4a | -0.121 |
| cx43.4 | 0.249 | ela2l | -0.121 |
| hmga1a | 0.247 | ela2 | -0.120 |
| marcksl1b | 0.246 | zgc:136461 | -0.119 |
| snrnp70 | 0.245 | mat1a | -0.119 |
| sox32 | 0.244 | ela3l | -0.119 |
| anp32e | 0.244 | prss59.1 | -0.118 |
| seta | 0.243 | spink4 | -0.118 |
| si:ch73-281n10.2 | 0.243 | apoa1b | -0.117 |
| stm | 0.242 | zgc:112160 | -0.116 |
| zmat2 | 0.241 | fbp1b | -0.116 |
| sp5l | 0.240 | zgc:158463 | -0.116 |
| zgc:110425 | 0.240 | sycn.2 | -0.115 |
| sox17 | 0.240 | cpb1 | -0.115 |
| safb | 0.238 | cpa5 | -0.115 |