striatin interacting protein 2
ZFIN 
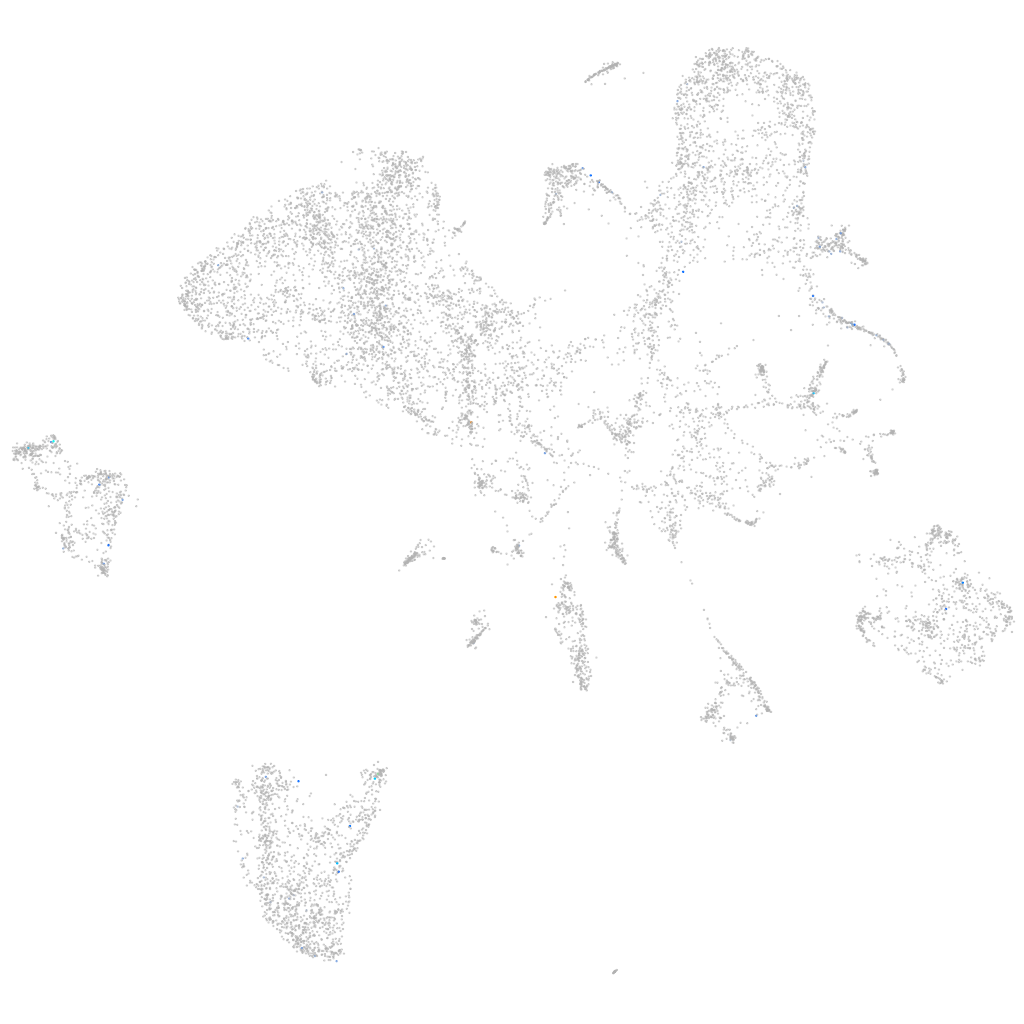
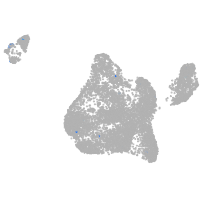
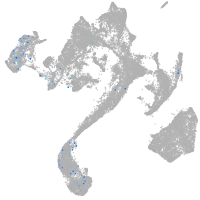
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-006820 | 0.433 | si:ch1073-325m22.2 | -0.042 |
| BX936415.1 | 0.429 | COX5B | -0.042 |
| trpc6b | 0.366 | cox5aa | -0.041 |
| gpr88 | 0.346 | hspe1 | -0.038 |
| BX001023.1 | 0.331 | cox7b | -0.035 |
| si:ch73-233f7.6 | 0.321 | rpl21 | -0.035 |
| zgc:153118 | 0.299 | cox7c | -0.034 |
| XLOC-015481 | 0.295 | cox4i1 | -0.033 |
| XLOC-036573 | 0.293 | apobb.1 | -0.033 |
| myadml2 | 0.287 | mdh1aa | -0.033 |
| frmpd3 | 0.279 | prdx2 | -0.033 |
| desmb | 0.261 | scp2a | -0.032 |
| LOC110439441 | 0.255 | serpina1l | -0.031 |
| lingo2a | 0.255 | atp5mc3a | -0.031 |
| kcnq3 | 0.253 | rgrb | -0.031 |
| LOC110438360 | 0.245 | gcshb | -0.031 |
| LOC101885553 | 0.243 | rack1 | -0.031 |
| prph2b | 0.235 | pcbd1 | -0.030 |
| XLOC-014755 | 0.229 | ftcd | -0.030 |
| aatka | 0.228 | atp5f1b | -0.030 |
| cacna1g | 0.224 | apom | -0.030 |
| nxph2b | 0.220 | pla2g12b | -0.030 |
| LOC110440168 | 0.212 | dbi | -0.030 |
| dnajb5 | 0.211 | gpd1b | -0.030 |
| CU861477.1 | 0.211 | atp5l | -0.030 |
| gabrd | 0.210 | cox6b2 | -0.029 |
| gdap1l1 | 0.205 | ndufs5 | -0.029 |
| BX510929.1 | 0.202 | pgrmc1 | -0.029 |
| LOC103908970 | 0.201 | pgam1a | -0.029 |
| mir132-2 | 0.199 | suclg2 | -0.029 |
| cacng2a | 0.193 | gstt1a | -0.029 |
| arhgap39 | 0.191 | cdo1 | -0.029 |
| gpr173 | 0.191 | si:ch73-361h17.1 | -0.028 |
| pou4f2 | 0.187 | fetub | -0.028 |
| fdx1b | 0.181 | slco1d1 | -0.028 |