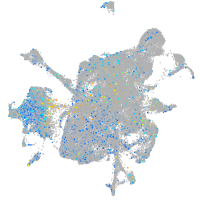
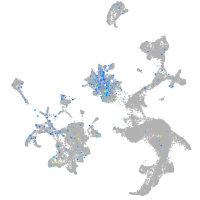
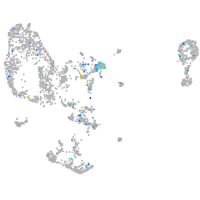
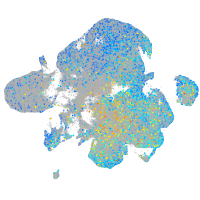
storkhead box 2b
ZFIN 
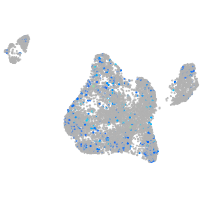
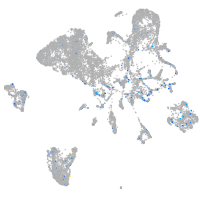
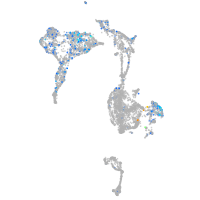
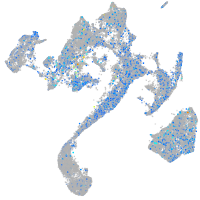
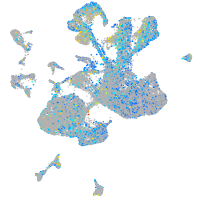
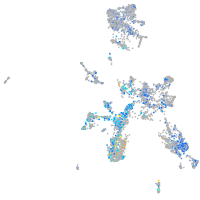
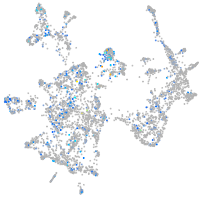
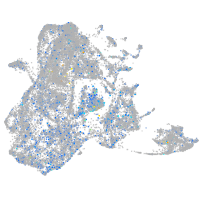
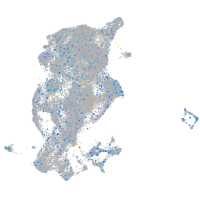
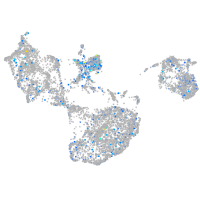
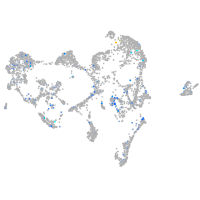
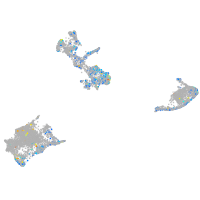
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108179280 | 0.183 | gapdh | -0.116 |
| LOC110438234 | 0.160 | aldob | -0.115 |
| epb41a | 0.155 | gamt | -0.110 |
| parp6a | 0.151 | fbp1b | -0.106 |
| ssx2ipb | 0.150 | scp2a | -0.101 |
| scrt2 | 0.150 | eno3 | -0.100 |
| NPFFR2 | 0.150 | ahcy | -0.099 |
| tmtops2b | 0.141 | gpx4a | -0.099 |
| tuba1c | 0.139 | suclg1 | -0.098 |
| CR392036.1 | 0.136 | gstt1a | -0.098 |
| tmeff1b | 0.136 | apoa4b.1 | -0.097 |
| BX936433.2 | 0.131 | apoa1b | -0.095 |
| chd5 | 0.130 | pgm1 | -0.093 |
| CABZ01118575.1 | 0.129 | apoc2 | -0.093 |
| myo9ab | 0.128 | glud1b | -0.092 |
| si:ch211-163m17.4 | 0.128 | pgk1 | -0.091 |
| nova2 | 0.127 | gatm | -0.091 |
| BX927388.1 | 0.126 | mat1a | -0.089 |
| tubb5 | 0.125 | pnp4b | -0.088 |
| hmgb3a | 0.124 | agxtb | -0.087 |
| jpt1b | 0.124 | pklr | -0.086 |
| smarce1 | 0.123 | agxta | -0.086 |
| bmf1 | 0.121 | sod1 | -0.085 |
| BX248128.2 | 0.120 | msrb2 | -0.085 |
| hcn4l | 0.120 | afp4 | -0.085 |
| calb2b | 0.119 | fabp10a | -0.084 |
| h3f3d | 0.119 | mgst1.2 | -0.084 |
| si:ch73-1a9.3 | 0.117 | ftcd | -0.084 |
| h2afva | 0.116 | grhprb | -0.083 |
| si:ch211-276a17.5 | 0.116 | mdh1aa | -0.083 |
| or111-5 | 0.115 | nipsnap3a | -0.083 |
| ubc | 0.115 | tdo2a | -0.083 |
| stap2a | 0.115 | cx32.3 | -0.083 |
| si:ch211-222l21.1 | 0.114 | gcshb | -0.082 |
| scrt1a | 0.114 | apom | -0.082 |