serine/threonine kinase 38 like
ZFIN 
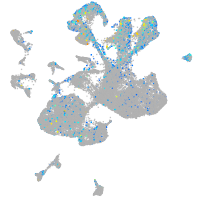
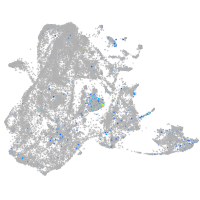
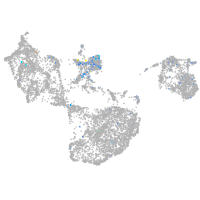
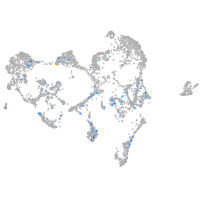
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.335 | gapdh | -0.118 |
| neurod1 | 0.307 | gamt | -0.117 |
| insm1b | 0.297 | gatm | -0.106 |
| pcsk1nl | 0.292 | mat1a | -0.102 |
| zgc:101731 | 0.285 | aqp12 | -0.100 |
| fev | 0.284 | ahcy | -0.100 |
| insm1a | 0.278 | nupr1b | -0.098 |
| egr4 | 0.278 | bhmt | -0.097 |
| mir7a-1 | 0.270 | cx32.3 | -0.096 |
| c2cd4a | 0.261 | agxtb | -0.095 |
| dll4 | 0.259 | hdlbpa | -0.095 |
| syt1a | 0.252 | fbp1b | -0.094 |
| mir375-2 | 0.248 | aldh6a1 | -0.094 |
| nkx2.2a | 0.244 | gstr | -0.090 |
| pax6b | 0.242 | aldh7a1 | -0.088 |
| pcsk1 | 0.241 | agxta | -0.087 |
| vat1 | 0.235 | apoc1 | -0.085 |
| scg2a | 0.233 | tdo2a | -0.085 |
| penka | 0.231 | aldob | -0.084 |
| si:dkey-153k10.9 | 0.230 | ttc36 | -0.084 |
| elovl1a | 0.229 | apoa4b.1 | -0.084 |
| calm1b | 0.229 | scp2a | -0.084 |
| vamp2 | 0.227 | nipsnap3a | -0.083 |
| capsla | 0.225 | rbp2b | -0.083 |
| sprn2 | 0.224 | mgst1.2 | -0.083 |
| scgn | 0.223 | cox7a1 | -0.082 |
| rgs8 | 0.222 | uox | -0.082 |
| rnasekb | 0.219 | apoa1b | -0.082 |
| XLOC-007078 | 0.219 | gstt1a | -0.082 |
| myt1b | 0.218 | fabp10a | -0.082 |
| gapdhs | 0.217 | rgrb | -0.081 |
| gpc1a | 0.216 | ttr | -0.081 |
| si:ch73-160i9.2 | 0.215 | phyhd1 | -0.080 |
| alpk3b | 0.215 | gnmt | -0.080 |
| tspan7b | 0.214 | hpda | -0.079 |