serine/threonine kinase 10
ZFIN 
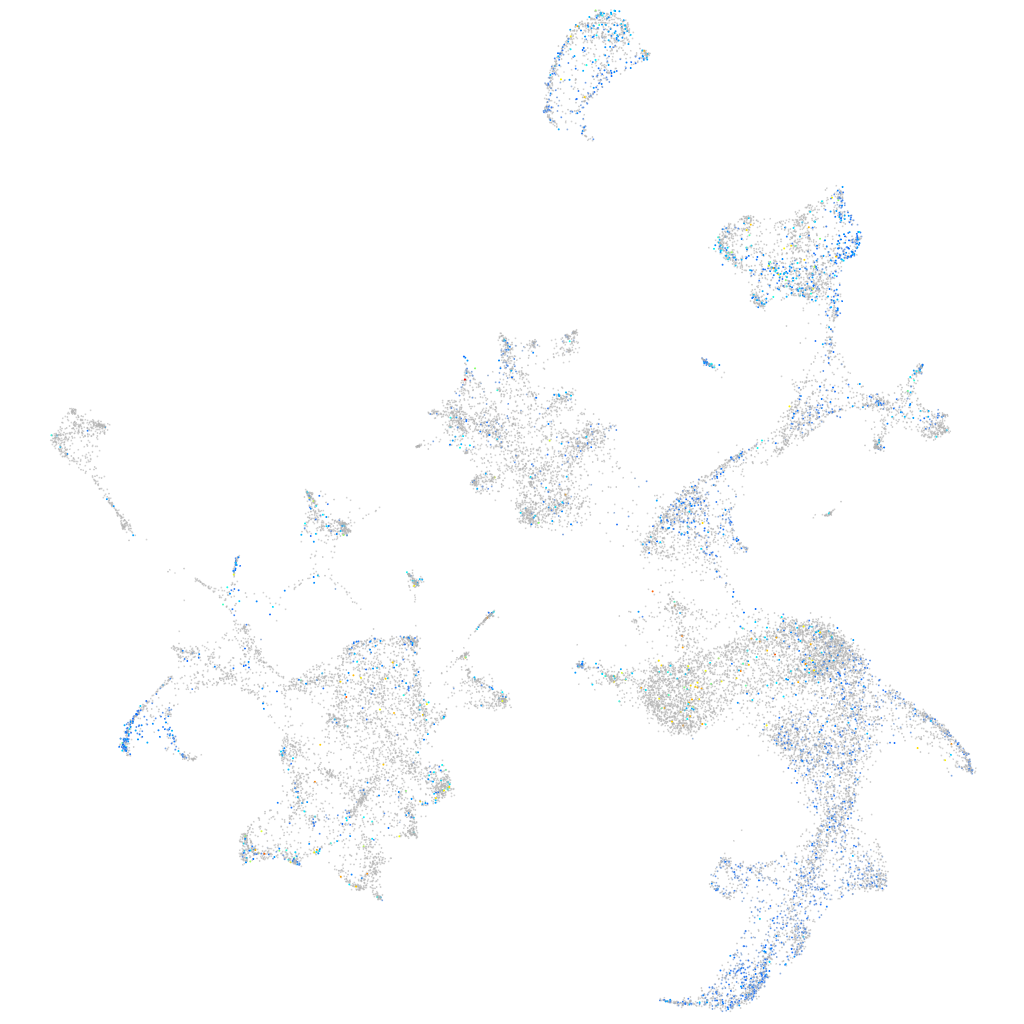
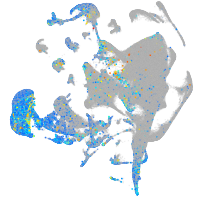
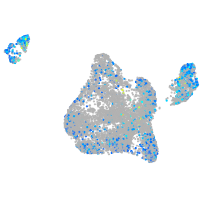
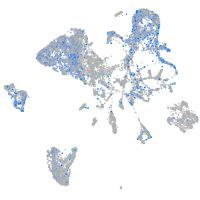
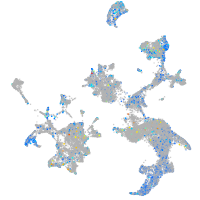
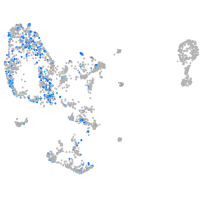
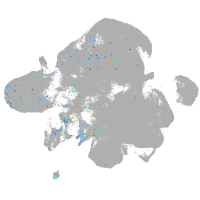
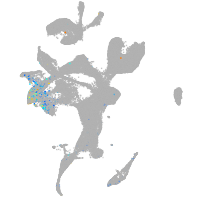
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| spi1b | 0.127 | hmgb3a | -0.064 |
| cebpa | 0.124 | marcksb | -0.061 |
| si:ch73-248e21.7 | 0.124 | hmgb1b | -0.058 |
| ptprc | 0.124 | tubb5 | -0.057 |
| ctss2.1 | 0.119 | tuba1a | -0.057 |
| nrros | 0.116 | mdkb | -0.056 |
| plxnc1 | 0.114 | jpt1b | -0.055 |
| il1b | 0.114 | marcksl1b | -0.054 |
| fybb | 0.113 | fscn1a | -0.054 |
| fcer1gl | 0.113 | pmp22a | -0.054 |
| laptm5 | 0.113 | tuba1c | -0.053 |
| ptpn6 | 0.112 | si:ch211-250g4.3 | -0.052 |
| rab44 | 0.112 | tmem88a | -0.049 |
| si:ch211-102c2.4 | 0.111 | elavl3 | -0.049 |
| cebpb | 0.109 | tmsb | -0.047 |
| si:ch211-212k18.7 | 0.108 | stmn1b | -0.047 |
| CABZ01041494.1 | 0.108 | nova2 | -0.047 |
| glipr1a | 0.107 | si:ch211-137a8.4 | -0.044 |
| mapre1a | 0.107 | rtn1a | -0.043 |
| txn | 0.107 | sox11a | -0.043 |
| arpc1b | 0.107 | zc4h2 | -0.043 |
| coro1a | 0.106 | tmeff1b | -0.042 |
| zgc:64051 | 0.105 | hbbe2 | -0.042 |
| cyba | 0.105 | si:ch73-46j18.5 | -0.042 |
| vsir | 0.105 | creg1 | -0.041 |
| myo1f | 0.105 | stxbp1a | -0.041 |
| marco | 0.105 | mdka | -0.040 |
| si:ch211-194m7.3 | 0.105 | gng3 | -0.040 |
| pgd | 0.105 | gpm6aa | -0.040 |
| CABZ01078737.1 | 0.104 | akap12b | -0.039 |
| spi1a | 0.102 | coro1cb | -0.039 |
| hcls1 | 0.102 | FO082781.1 | -0.039 |
| rac2 | 0.101 | dpysl3 | -0.039 |
| rnaseka | 0.101 | etv2 | -0.039 |
| grna | 0.101 | tmem88b | -0.039 |