"ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2"
ZFIN 
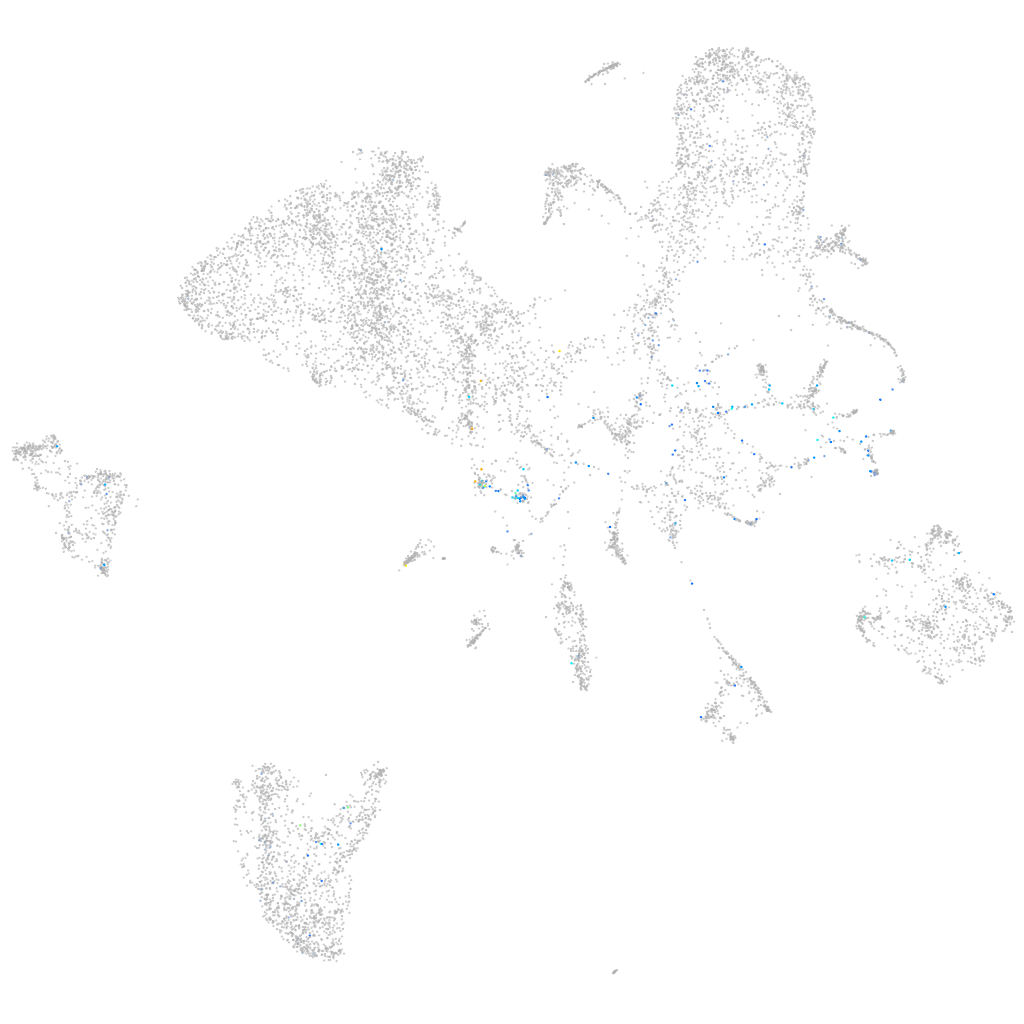
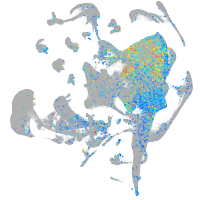
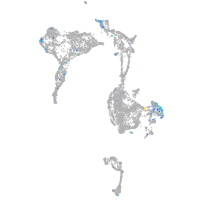
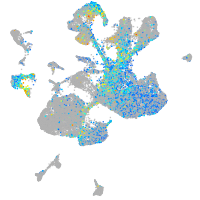
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kiaa1549lb | 0.197 | eno3 | -0.089 |
| LOC100332354 | 0.195 | aldob | -0.088 |
| LOC798783 | 0.190 | gapdh | -0.086 |
| myt1a | 0.186 | scp2a | -0.085 |
| elavl3 | 0.185 | glud1b | -0.082 |
| nova2 | 0.182 | fbp1b | -0.080 |
| XLOC-003689 | 0.177 | pgk1 | -0.078 |
| abhd6a | 0.175 | gpx4a | -0.078 |
| tmeff1b | 0.171 | ckba | -0.076 |
| tuba1c | 0.167 | cx32.3 | -0.075 |
| myo16 | 0.166 | mgst1.2 | -0.075 |
| fam171b | 0.165 | aldh7a1 | -0.074 |
| CT573433.1 | 0.163 | sod1 | -0.073 |
| LOC101883911 | 0.163 | apoa4b.1 | -0.073 |
| her4.1 | 0.160 | gcshb | -0.072 |
| erbb4a | 0.160 | gamt | -0.072 |
| si:ch211-57n23.4 | 0.158 | pnp4b | -0.072 |
| XLOC-002952 | 0.158 | agxtb | -0.072 |
| srgap3 | 0.158 | gnmt | -0.071 |
| l1cama | 0.153 | apoc2 | -0.071 |
| fam163b | 0.153 | pgm1 | -0.070 |
| pcdh2ab7 | 0.153 | abat | -0.070 |
| pax3a | 0.151 | gatm | -0.070 |
| her4.2 | 0.150 | isoc2 | -0.070 |
| tubb5 | 0.149 | msrb2 | -0.069 |
| XLOC-003690 | 0.148 | hpda | -0.069 |
| mir219-3 | 0.148 | suclg1 | -0.069 |
| myt1la | 0.147 | upb1 | -0.069 |
| rtn1b | 0.146 | ttc36 | -0.068 |
| her4.4 | 0.146 | uox | -0.068 |
| LOC110440219 | 0.144 | aldh6a1 | -0.068 |
| htr7c | 0.144 | etnppl | -0.068 |
| rai1 | 0.144 | fdx1 | -0.068 |
| si:ch73-21g5.7 | 0.144 | fabp10a | -0.068 |
| AL954831.1 | 0.144 | mat1a | -0.068 |