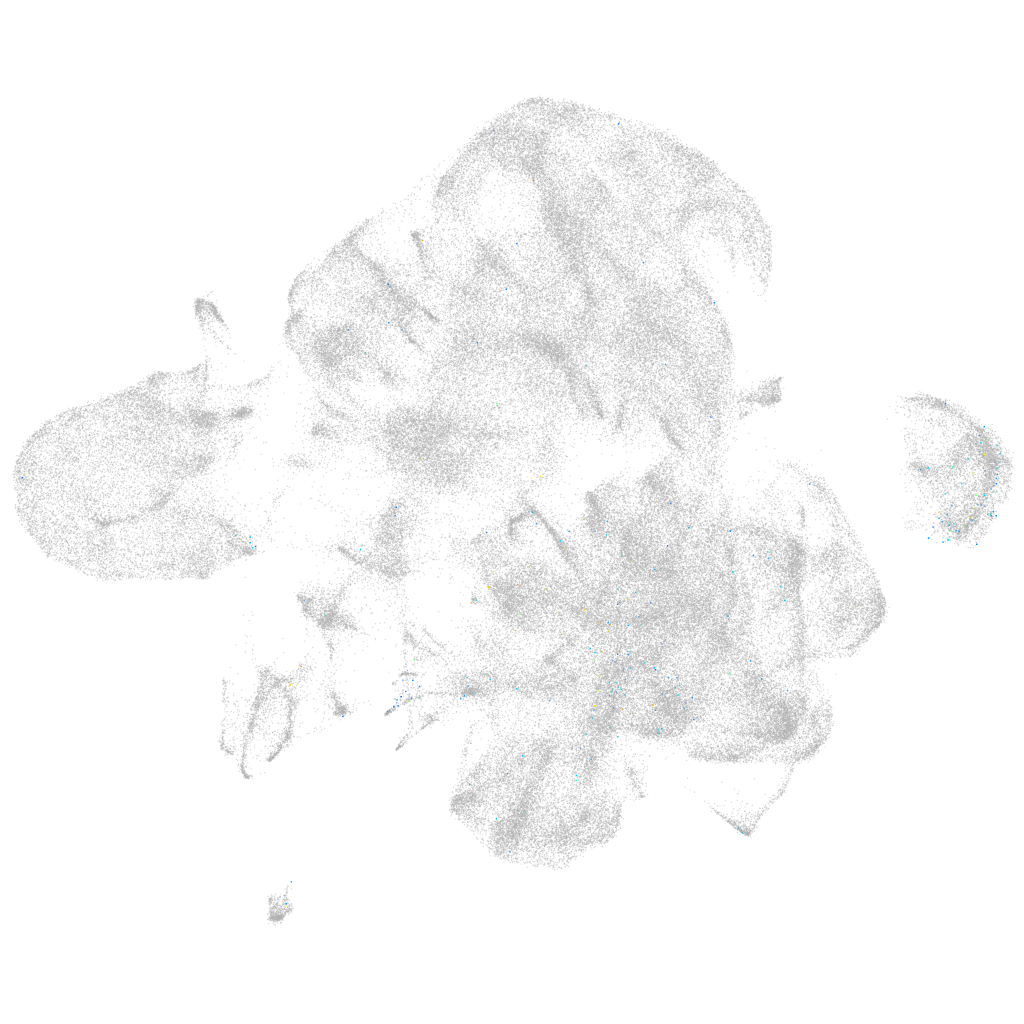
somatostatin receptor 5
ZFIN 
Other cell groups


all cells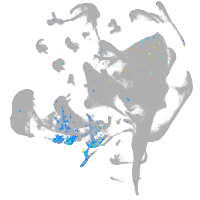 |
blastomeres |
PGCs |
endoderm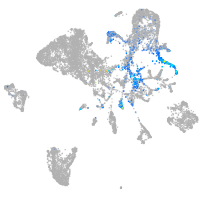 |
axial mesoderm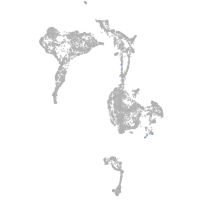 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line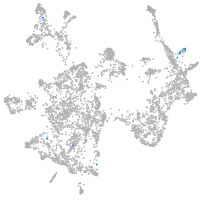 |
epidermis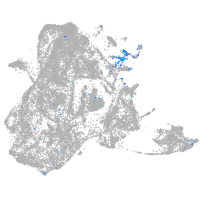 |
periderm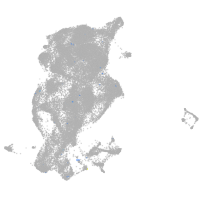 |
fin |
ionocytes / mucous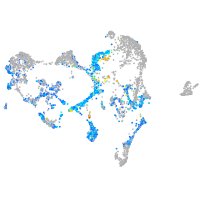 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101886574 | 0.172 | hmgb2b | -0.037 |
| LO018333.1 | 0.096 | rpl11 | -0.035 |
| spx | 0.086 | rps27a | -0.034 |
| prop1 | 0.080 | ran | -0.033 |
| c19h1orf216 | 0.072 | eef1a1l1 | -0.033 |
| ednrbb | 0.070 | hmga1a | -0.033 |
| XLOC-020906 | 0.070 | khdrbs1a | -0.032 |
| BX248331.2 | 0.067 | rplp1 | -0.032 |
| acsl3a | 0.061 | rps2 | -0.032 |
| si:rp71-1c10.8 | 0.055 | rps4x | -0.031 |
| LOC110438412 | 0.055 | si:dkey-151g10.6 | -0.031 |
| dre-let-7a-1 | 0.053 | rps24 | -0.031 |
| foxi3b | 0.053 | rpl7 | -0.031 |
| synpr | 0.052 | rps20 | -0.030 |
| c16h2orf66 | 0.048 | rpl15 | -0.029 |
| glis1a | 0.048 | snrpf | -0.029 |
| CU468723.1 | 0.047 | rpl7a | -0.029 |
| si:dkey-192g7.3 | 0.047 | rps12 | -0.029 |
| LO017820.1 | 0.046 | rpl10a | -0.029 |
| lepr | 0.046 | rack1 | -0.028 |
| cnfn | 0.046 | rpsa | -0.028 |
| cfap300 | 0.044 | rpl17 | -0.028 |
| scg2b | 0.044 | rpl35a | -0.028 |
| npffr2b | 0.044 | rps11 | -0.028 |
| XLOC-044140 | 0.044 | rps25 | -0.028 |
| cremb | 0.043 | rpl19 | -0.028 |
| BX927335.2 | 0.043 | rps7 | -0.028 |
| sync | 0.042 | rpl8 | -0.028 |
| slc32a1 | 0.041 | rps19 | -0.028 |
| gad2 | 0.040 | hmgb2a | -0.028 |
| egr4 | 0.040 | rps13 | -0.028 |
| gpa33b | 0.040 | rpl27 | -0.028 |
| atp6ap2 | 0.040 | ybx1 | -0.027 |
| htra1b | 0.040 | syncrip | -0.027 |
| dlx5a | 0.040 | rpl23 | -0.027 |