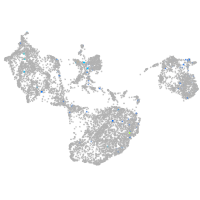
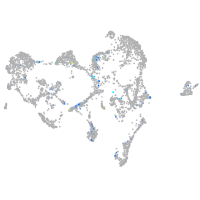
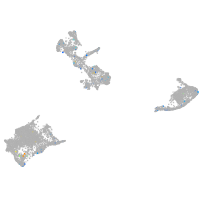
slingshot protein phosphatase 1a
ZFIN 
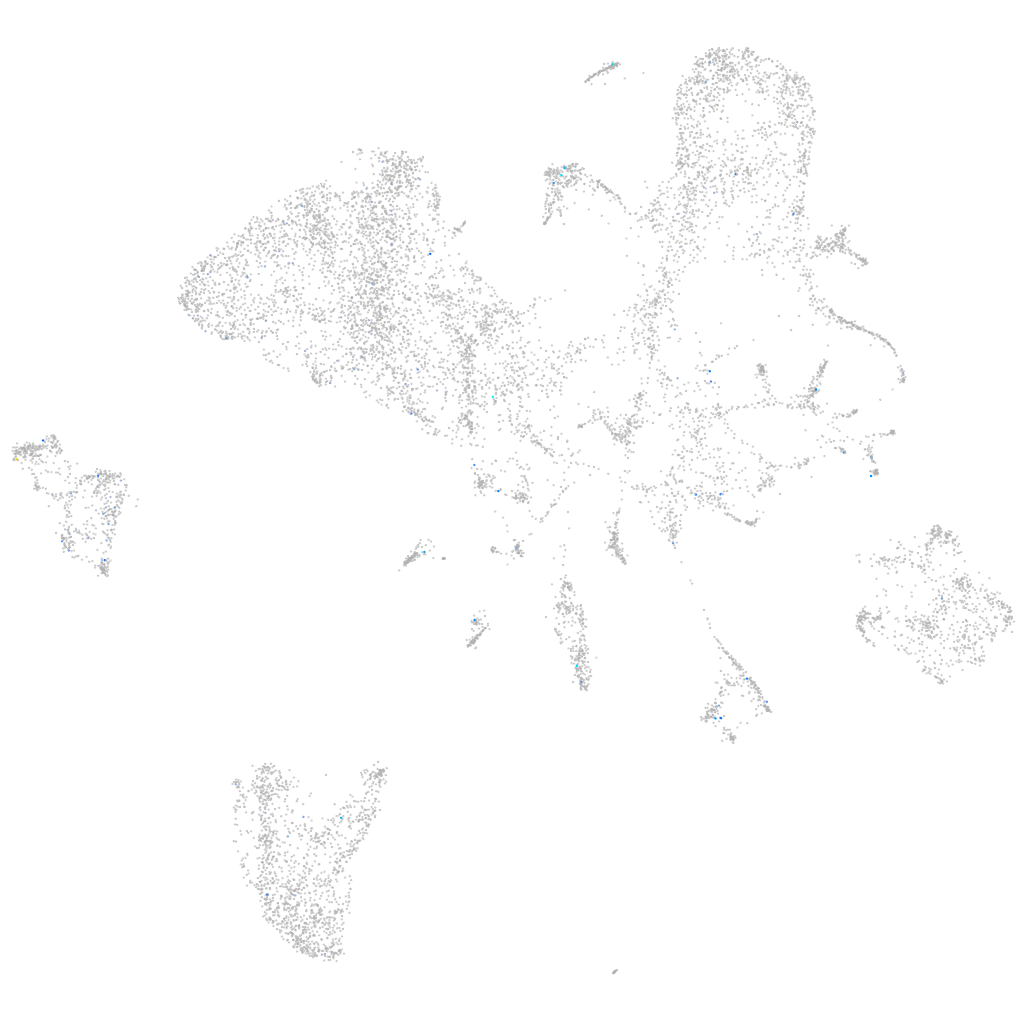
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101887092 | 0.248 | gapdh | -0.028 |
| BX936308.2 | 0.209 | cnbpa | -0.026 |
| FP325124.1 | 0.203 | eif5b | -0.026 |
| AL954142.2 | 0.192 | cldn15lb | -0.026 |
| CU651662.1 | 0.177 | cycsb | -0.024 |
| LOC101885980 | 0.165 | mat1a | -0.023 |
| nitr8 | 0.160 | bhmt | -0.023 |
| CABZ01046980.1 | 0.148 | ttpa | -0.023 |
| nkl.2 | 0.145 | ahcy | -0.023 |
| LOC101886291 | 0.141 | eif4e1c | -0.023 |
| si:ch73-44m9.3 | 0.138 | hspe1 | -0.022 |
| si:dkey-22i16.9 | 0.137 | agxtb | -0.022 |
| AL954361.1 | 0.130 | cox7a1 | -0.022 |
| capn3a | 0.122 | ptges3b | -0.022 |
| tecta | 0.120 | si:dkey-16p21.8 | -0.022 |
| XLOC-040894 | 0.118 | pnp4b | -0.022 |
| XLOC-042485 | 0.118 | aqp12 | -0.021 |
| pou4f2 | 0.112 | gstt1a | -0.021 |
| nppa | 0.110 | phyhd1 | -0.021 |
| pmelb | 0.110 | gamt | -0.021 |
| map3k8 | 0.107 | ilf2 | -0.021 |
| CU550702.1 | 0.105 | ckma | -0.021 |
| CR376774.1 | 0.105 | scp2a | -0.021 |
| CU672266.1 | 0.104 | chchd10 | -0.020 |
| dclk2b | 0.101 | gnmt | -0.020 |
| csf1rb | 0.100 | bms1 | -0.020 |
| kcna6a | 0.099 | zmp:0000000624 | -0.020 |
| LOC110439528 | 0.099 | agmo | -0.020 |
| BX005351.1 | 0.098 | eno3 | -0.020 |
| slc6a6a | 0.097 | smarcd1 | -0.020 |
| lrch2 | 0.097 | gatm | -0.020 |
| scfd2 | 0.096 | mdh1aa | -0.020 |
| LOC103908707 | 0.095 | cth | -0.020 |
| BX248511.1 | 0.095 | ftcd | -0.020 |
| kank1b | 0.093 | esf1 | -0.020 |