serine/arginine repetitive matrix 2
ZFIN 
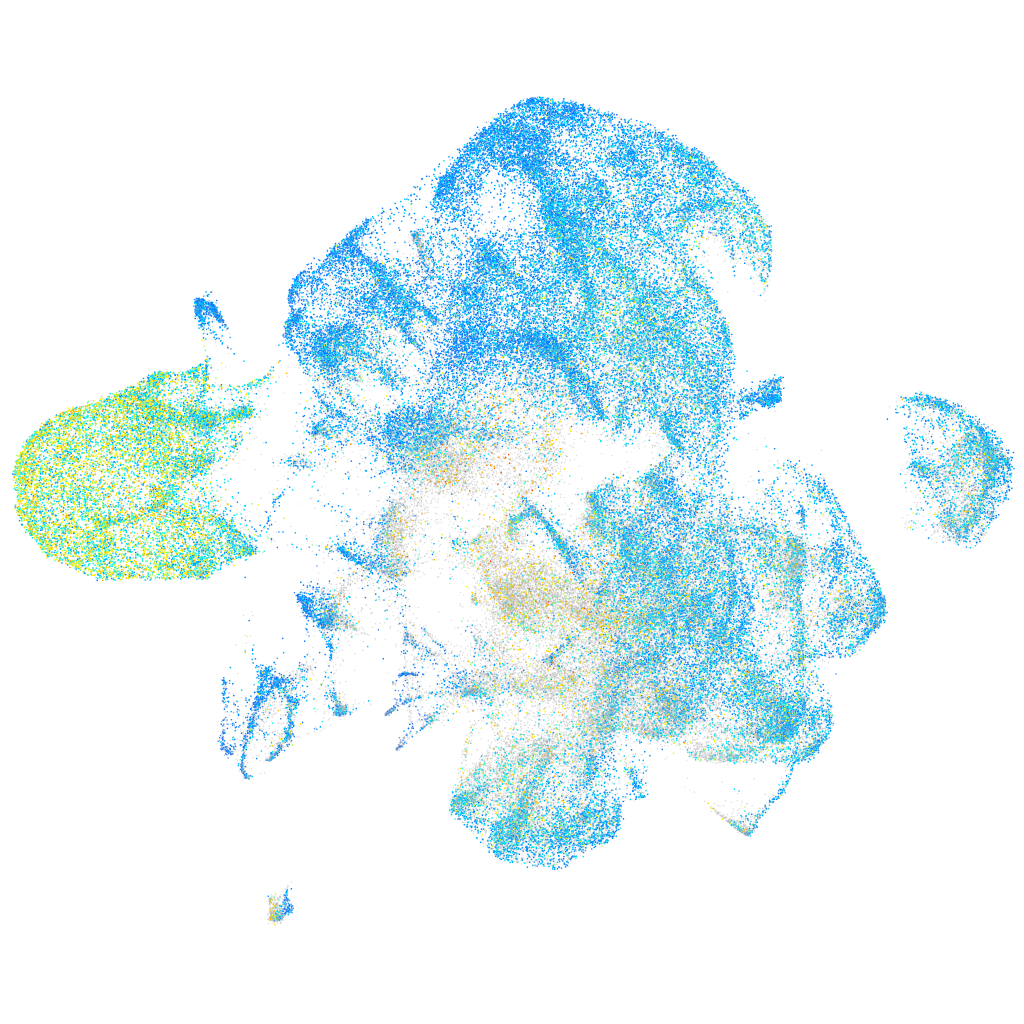
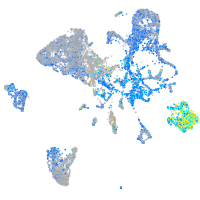
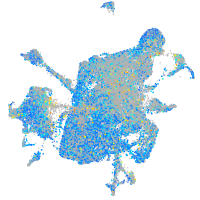
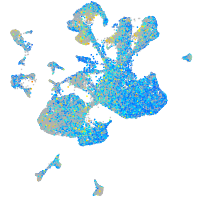
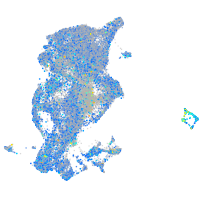
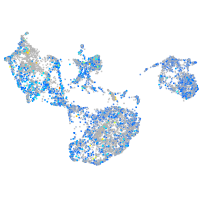
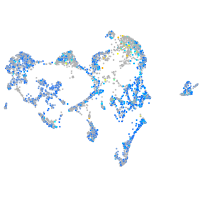
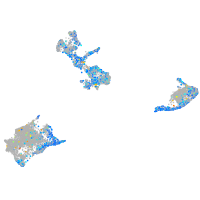
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-152c2.3 | 0.403 | ptmaa | -0.278 |
| hspb1 | 0.396 | rpl37 | -0.258 |
| stm | 0.377 | rps10 | -0.238 |
| fbl | 0.362 | CR383676.1 | -0.231 |
| nop58 | 0.360 | zgc:114188 | -0.222 |
| dkc1 | 0.355 | rtn1a | -0.209 |
| apoeb | 0.352 | pvalb1 | -0.208 |
| ncl | 0.350 | gpm6aa | -0.205 |
| npm1a | 0.350 | actc1b | -0.204 |
| hnrnpa1b | 0.344 | pvalb2 | -0.203 |
| nop56 | 0.344 | tuba1c | -0.196 |
| NC-002333.4 | 0.338 | tmsb4x | -0.196 |
| pou5f3 | 0.335 | stmn1b | -0.193 |
| polr3gla | 0.334 | gpm6ab | -0.190 |
| crabp2b | 0.333 | h3f3a | -0.190 |
| nop2 | 0.332 | atp6v0cb | -0.186 |
| anp32e | 0.329 | rnasekb | -0.183 |
| zmp:0000000624 | 0.329 | zgc:158463 | -0.178 |
| bms1 | 0.324 | ppiab | -0.174 |
| gnl3 | 0.324 | COX3 | -0.172 |
| lig1 | 0.322 | ppdpfb | -0.171 |
| ppig | 0.321 | elavl3 | -0.169 |
| gar1 | 0.321 | mt-atp6 | -0.168 |
| si:dkey-66i24.9 | 0.319 | sncb | -0.168 |
| s100a1 | 0.315 | calm1a | -0.166 |
| akap12b | 0.314 | gapdhs | -0.165 |
| rsl1d1 | 0.314 | marcksl1a | -0.163 |
| ebna1bp2 | 0.314 | mt-nd1 | -0.159 |
| apoc1 | 0.311 | ckbb | -0.159 |
| hmga1a | 0.310 | gng3 | -0.156 |
| ddx18 | 0.309 | rtn1b | -0.155 |
| apela | 0.308 | vamp2 | -0.153 |
| kri1 | 0.305 | hbbe1.3 | -0.152 |
| nr6a1a | 0.302 | elavl4 | -0.152 |
| seta | 0.300 | hbae3 | -0.151 |