sterol regulatory element binding transcription factor 1
ZFIN 
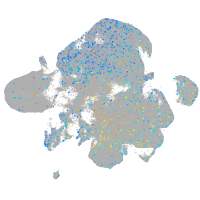
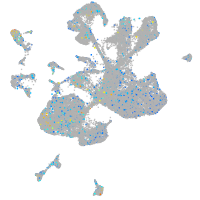
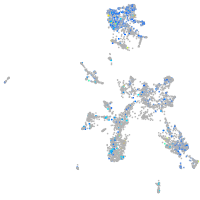
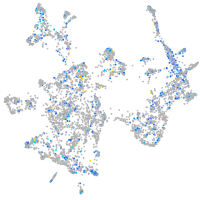
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ckbb | 0.055 | rps24 | -0.046 |
| LOC108179355 | 0.053 | rplp1 | -0.046 |
| gpm6aa | 0.052 | rps23 | -0.044 |
| CU929317.1 | 0.051 | rpl26 | -0.043 |
| XLOC-000324 | 0.050 | rps12 | -0.042 |
| vcam1a | 0.050 | rps10 | -0.042 |
| mt-atp6 | 0.049 | faua | -0.039 |
| pkma | 0.047 | rpl27a | -0.039 |
| zgc:165518 | 0.047 | rps20 | -0.038 |
| tuba1c | 0.046 | rpl23 | -0.038 |
| rnasekb | 0.046 | si:dkey-151g10.6 | -0.038 |
| sypb | 0.045 | rps19 | -0.037 |
| gapdhs | 0.045 | rpl39 | -0.037 |
| mt-nd4 | 0.044 | rpl13a | -0.037 |
| mt-cyb | 0.044 | rps15 | -0.036 |
| LOC108183756 | 0.044 | rpl7 | -0.036 |
| atp1a1a.1 | 0.043 | eef1b2 | -0.036 |
| atp6v0cb | 0.043 | rpl13 | -0.036 |
| slc7a2 | 0.043 | rpl35a | -0.036 |
| LOC101886073 | 0.043 | rpl9 | -0.035 |
| pacsin1a | 0.042 | rps2 | -0.035 |
| elmo1 | 0.042 | rpl10 | -0.035 |
| XLOC-038795 | 0.042 | rps27a | -0.035 |
| atp1b2a | 0.042 | rps9 | -0.035 |
| olfm1b | 0.042 | rpl28 | -0.035 |
| CU856539.1 | 0.042 | rpl27 | -0.035 |
| gnao1b | 0.041 | rpl32 | -0.035 |
| zgc:158423 | 0.041 | rps3a | -0.035 |
| slc38a2 | 0.041 | rps21 | -0.035 |
| nova2 | 0.041 | rpl17 | -0.035 |
| fabp11a | 0.041 | rplp0 | -0.034 |
| nsg2 | 0.041 | rps13 | -0.034 |
| slc7a5 | 0.041 | rps26 | -0.034 |
| scarb2a | 0.041 | rpl36a | -0.034 |
| si:ch211-129p13.1 | 0.040 | rpl19 | -0.034 |