"steroid-5-alpha-reductase, alpha polypeptide 2b"
ZFIN 
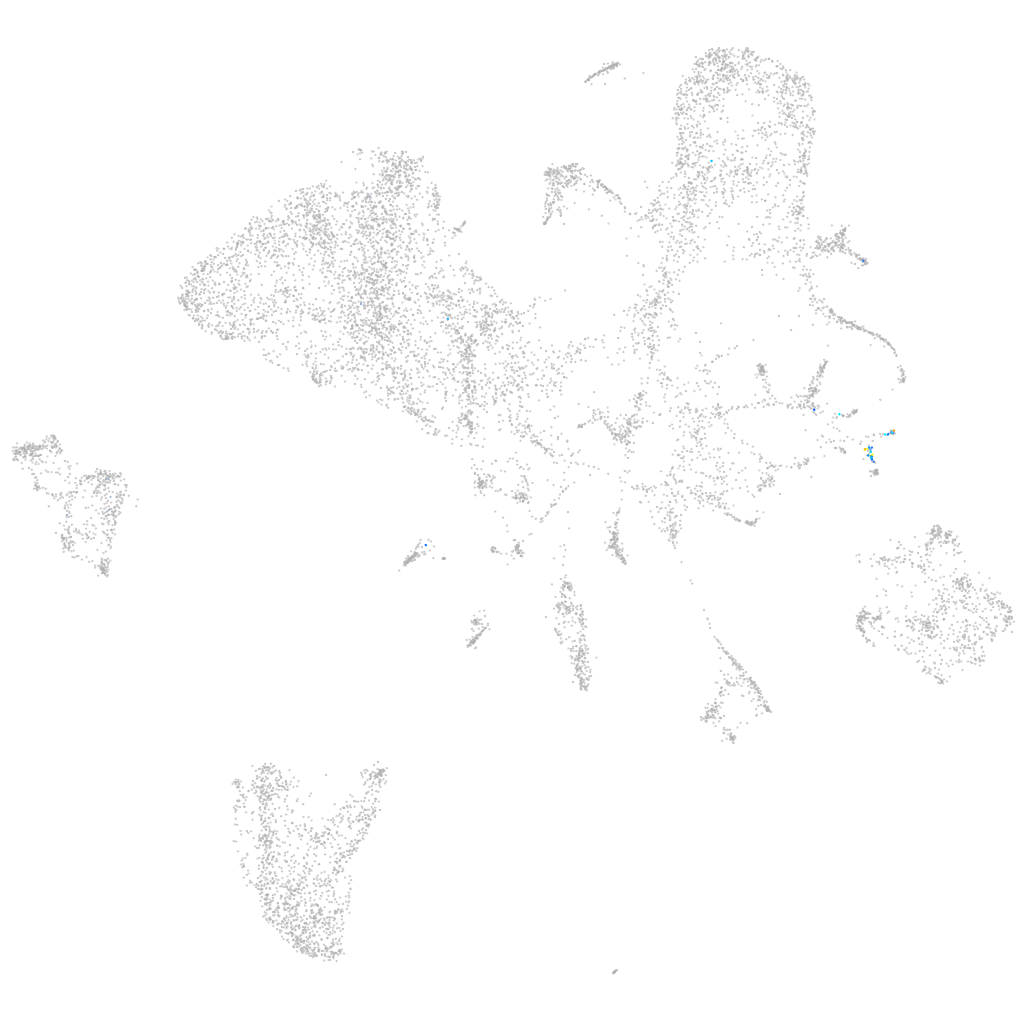
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm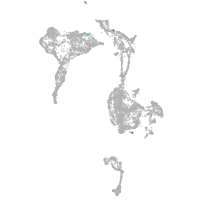 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 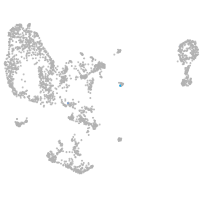 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis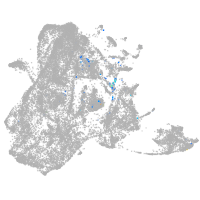 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sst1.1 | 0.521 | ahcy | -0.054 |
| BX571707.1 | 0.431 | aldob | -0.053 |
| sst1.2 | 0.345 | gapdh | -0.047 |
| ucn3l | 0.344 | gamt | -0.044 |
| gnrh2 | 0.317 | rps20 | -0.042 |
| tmem163b | 0.316 | aldh9a1a.1 | -0.041 |
| dkk3b | 0.308 | gstp1 | -0.041 |
| SLC5A10 | 0.292 | nupr1b | -0.041 |
| si:dkey-183i3.9 | 0.276 | cox6b1 | -0.041 |
| LOC571123 | 0.273 | gstr | -0.041 |
| pdyn | 0.266 | glud1b | -0.041 |
| casr | 0.265 | fbp1b | -0.040 |
| slitrk1 | 0.264 | gatm | -0.040 |
| cpne4a | 0.262 | bhmt | -0.040 |
| kcnc4 | 0.262 | hdlbpa | -0.039 |
| CT583625.5 | 0.260 | uqcrc2a | -0.038 |
| sst2 | 0.259 | tuba8l2 | -0.037 |
| GRIK2 | 0.255 | fabp3 | -0.036 |
| hhla2a.2 | 0.252 | aqp12 | -0.036 |
| scgn | 0.251 | mat1a | -0.036 |
| mtcl1 | 0.250 | si:dkey-16p21.8 | -0.036 |
| cadm2a | 0.248 | eno3 | -0.036 |
| kcnip3b | 0.246 | lgals2b | -0.035 |
| nobox | 0.242 | rmdn1 | -0.035 |
| hhex | 0.233 | agxta | -0.035 |
| gpc1a | 0.229 | afp4 | -0.035 |
| gpr153 | 0.224 | apoc2 | -0.034 |
| LOC110438434 | 0.221 | apoa4b.1 | -0.034 |
| ndufa4l2a | 0.219 | tktb | -0.034 |
| si:dkey-153k10.9 | 0.218 | tbca | -0.034 |
| kcnt1 | 0.216 | atp1a1a.1 | -0.034 |
| pcdh15b | 0.214 | cox8b | -0.034 |
| eif3bb | 0.214 | CABZ01032488.1 | -0.034 |
| rprmb | 0.210 | pa2g4a | -0.034 |
| scg2a | 0.204 | pa2g4b | -0.034 |